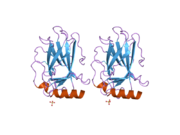- p53
-
For the band and album of the same name, see P53 (band) and P53 (album).
p53 (also known as protein 53 or tumor protein 53), is a tumor suppressor protein that in humans is encoded by the TP53 gene.[1][2][3][4] p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer. As such, p53 has been described as "the guardian of the genome", the "guardian angel gene", and the "master watchman", referring to its role in conserving stability by preventing genome mutation.[5]
The name p53 is in reference to its apparent molecular mass: It runs as a 53-kilodalton (kDa) protein on SDS-PAGE. But, based on calculations from its amino acid residues, p53's mass is actually only 43.7 kDa. This difference is due to the high number of proline residues in the protein, which slow its migration on SDS-PAGE, thus making it appear heavier than it actually is.[6] This effect is observed with p53 from a variety of species, including humans, rodents, frogs, and fish.
Contents
Nomenclature
p53 is also known as:
- UniProt name: Cellular tumor antigen p53
- Antigen NY-CO-13
- Phosphoprotein p53
- Transformation-related protein 53 (TRP53)
- Tumor suppressor p53
Gene
In humans, p53 is encoded by the TP53 gene located on the short arm of chromosome 17 (17p13.1).[1][2][3][4] The gene spans 20 kb, with a non-coding exon 1 and a very long first intron of 10 kb.The coding sequence contains five regions showing a high degree of conservation in vertebrates, predominantly in exons 2, 5, 6, 7 and 8, but the sequences found in invertebrates show only distant resemblance to mammalian TP53.[7] TP53 orthologs [8] have been identified in most mammals for which complete genome data are available.
In humans, a common polymorphism involves the substitution of an arginine for a proline at codon position 72. Many studies have investigated a genetic link between this variation and cancer susceptibility, however, a combined analysis did not show a link.[9]
For these mammals, the gene is located on different chromosomes:
- Chimp and orangutan, chromosome 17
- Macaque, chromosome 16
- Mouse, chromosome 11
- Rat, chromosome 10
- Dog, chromosome 5
- Cow, chromosome 19
- Pig, chromosome 12
- Horse, chromosome 11
- Opossum, chromosome 2
(Italics are used to denote the TP53 gene name and distinguish it from the protein it encodes.)
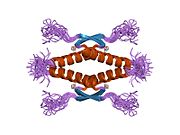
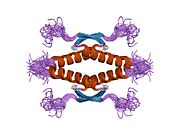
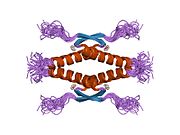
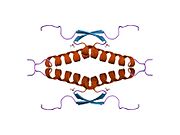
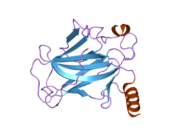
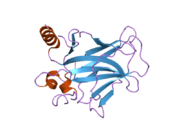
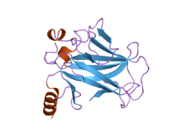
Structure
Human p53 is 393 amino acids long and has seven domains:
- an acidic N-terminus transcription-activation domain (TAD), also known as activation domain 1 (AD1), which activates transcription factors: residues 1-42. The N-terminus contains two complementary transcriptional activation domains, with a major one at residues 1–42 and a minor one at residues 55–75, specifically involved in the regulation of several pro-apoptotic genes.[10]
- activation domain 2 (AD2) important for apoptotic activity: residues 43-63.
- Proline rich domain important for the apoptotic activity of p53: residues 64-92.
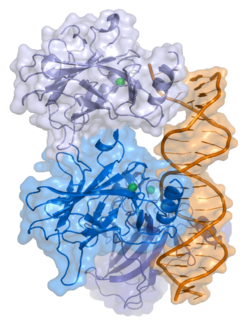
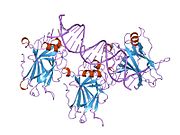
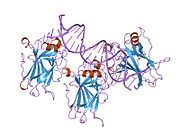
- central DNA-binding core domain (DBD). Contains one zinc atom and several arginine amino acids: residues 102-292. This region is responsible for binding the p53 co-repressor LMO3.[11]
- nuclear localization signaling domain, residues 316-325.
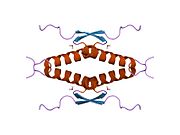
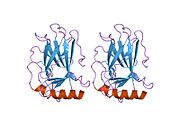
- homo-oligomerisation domain (OD): residues 307-355. Tetramerization is essential for the activity of p53 in vivo.
- C-terminal involved in downregulation of DNA binding of the central domain: residues 356-393.[12]
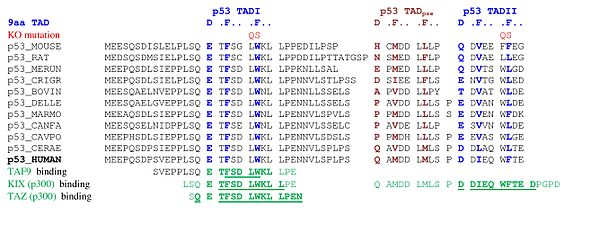
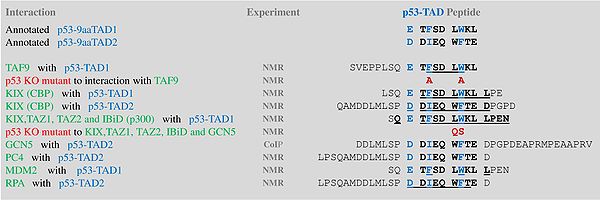
A tandem of nine-amino-acid transactivation domains (9aaTAD) was identified in the AD1 and AD2 regions of transcription factor p53.[13] KO mutations and position for p53 interaction with TFIID are listed below:[14]
9aaTADs mediate p53 interaction with general coactivators - TAF9, CBP/p300 (all four domains KIX, TAZ1, TAZ2 and IBiD), GCN5 and PC4, regulatory protein MDM2 and replication protein A (RPA).[15][16]
Mutations that deactivate p53 in cancer usually occur in the DBD. Most of these mutations destroy the ability of the protein to bind to its target DNA sequences, and thus prevents transcriptional activation of these genes. As such, mutations in the DBD are recessive loss-of-function mutations. Molecules of p53 with mutations in the OD dimerise with wild-type p53, and prevent them from activating transcription. Therefore OD mutations have a dominant negative effect on the function of p53.
Wild-type p53 is a labile protein, comprising folded and unstructured regions that function in a synergistic manner.[17]
Function
p53 has many mechanisms of anticancer function, and plays a role in apoptosis, genomic stability, and inhibition of angiogenesis. In its anti-cancer role, p53 works through several mechanisms:
- It can activate DNA repair proteins when DNA has sustained damage.
- It can induce growth arrest by holding the cell cycle at the G1/S regulation point on DNA damage recognition (if it holds the cell here for long enough, the DNA repair proteins will have time to fix the damage and the cell will be allowed to continue the cell cycle).
- It can initiate apoptosis, the programmed cell death, if DNA damage proves to be irreparable.
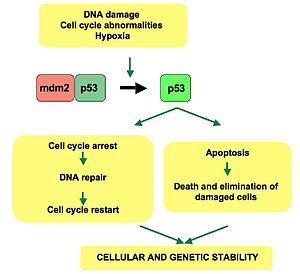 p53 pathway: In a normal cell p53 is inactivated by its negative regulator, mdm2. Upon DNA damage or other stresses, various pathways will lead to the dissociation of the p53 and mdm2 complex. Once activated, p53 will induce a cell cycle arrest to allow either repair and survival of the cell or apoptosis to discard the damaged cell. How p53 makes this choice is currently unknown.
p53 pathway: In a normal cell p53 is inactivated by its negative regulator, mdm2. Upon DNA damage or other stresses, various pathways will lead to the dissociation of the p53 and mdm2 complex. Once activated, p53 will induce a cell cycle arrest to allow either repair and survival of the cell or apoptosis to discard the damaged cell. How p53 makes this choice is currently unknown.
Activated p53 binds DNA and activates expression of several genes including WAF1/CIP1 encoding for p21. p21 (WAF1) binds to the G1-S/CDK (CDK2) and S/CDK complexes (molecules important for the G1/S transition in the cell cycle) inhibiting their activity.
When p21(WAF1) is complexed with CDK2 the cell cannot continue to the next stage of cell division. A mutant p53 will no longer bind DNA in an effective way, and, as a consequence, the p21 protein will not be available to act as the "stop signal" for cell division. Thus, cells will divide uncontrollably, and form tumors.[18]
Recent research has also linked the p53 and RB1 pathways, via p14ARF, raising the possibility that the pathways may regulate each other.[19]
p53 by regulating LIF has been shown to facilitate implantation in the mouse model and possibly in humans.[20]
p53 expression can be stimulated by UV light, which also causes DNA damage. In this case, p53 can initiate events leading to tanning.[21][22]
Regulation
p53 becomes activated in response to a myriad of stress types, which include but are not limited to DNA damage (induced by either UV, IR, or chemical agents such as hydrogen peroxide), oxidative stress,[23] osmotic shock, ribonucleotide depletion, and deregulated oncogene expression. This activation is marked by two major events. First, the half-life of the p53 protein is increased drastically, leading to a quick accumulation of p53 in stressed cells. Second, a conformational change forces p53 to be activated as a transcription regulator in these cells. The critical event leading to the activation of p53 is the phosphorylation of its N-terminal domain. The N-terminal transcriptional activation domain contains a large number of phosphorylation sites and can be considered as the primary target for protein kinases transducing stress signals.
The protein kinases that are known to target this transcriptional activation domain of p53 can be roughly divided into two groups. A first group of protein kinases belongs to the MAPK family (JNK1-3, ERK1-2, p38 MAPK), which is known to respond to several types of stress, such as membrane damage, oxidative stress, osmotic shock, heat shock, etc. A second group of protein kinases (ATR, ATM, CHK1 and CHK2, DNA-PK, CAK) is implicated in the genome integrity checkpoint, a molecular cascade that detects and responds to several forms of DNA damage caused by genotoxic stress. Oncogenes also stimulate p53 activation, mediated by the protein p14ARF.
In unstressed cells, p53 levels are kept low through a continuous degradation of p53. A protein called Mdm2 (also called HDM2 in humans), which is itself a product of p53, binds to p53, preventing its action and transports it from the nucleus to the cytosol. Also Mdm2 acts as ubiquitin ligase and covalently attaches ubiquitin to p53 and thus marks p53 for degradation by the proteasome. However, ubiquitylation of p53 is reversible. A ubiquitin specific protease, USP7 (or HAUSP), can cleave ubiquitin off p53, thereby protecting it from proteasome-dependent degradation. This is one means by which p53 is stabilized in response to oncogenic insults.
Phosphorylation of the N-terminal end of p53 by the above-mentioned protein kinases disrupts Mdm2-binding. Other proteins, such as Pin1, are then recruited to p53 and induce a conformational change in p53, which prevents Mdm2-binding even more. Phosphorylation also allows for binding of transcriptional coactivators, like p300 or PCAF, which then acetylate the carboxy-terminal end of p53, exposing the DNA binding domain of p53, allowing it to activate or repress specific genes. Deacetylase enzymes, such as Sirt1 and Sirt7, can deacetylate p53, leading to an inhibition of apoptosis.[24] Some oncogenes can also stimulate the transcription of proteins that bind to MDM2 and inhibit its activity.
Role in disease
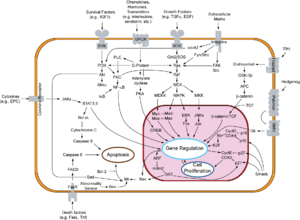 Overview of signal transduction pathways involved in apoptosis.
Overview of signal transduction pathways involved in apoptosis.
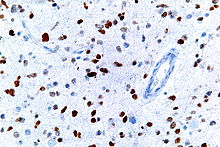 A micrograph showing cells with abnormal p53 expression (brown) in a brain tumour. p53 immunostain.
A micrograph showing cells with abnormal p53 expression (brown) in a brain tumour. p53 immunostain.
If the TP53 gene is damaged, tumor suppression is severely reduced. People who inherit only one functional copy of the TP53 gene will most likely develop tumors in early adulthood, a disease known as Li-Fraumeni syndrome. The TP53 gene can also be damaged in cells by mutagens (chemicals, radiation, or viruses), increasing the likelihood that the cell will begin decontrolled division. More than 50 percent of human tumors contain a mutation or deletion of the TP53 gene.[25] Increasing the amount of p53, which may initially seem a good way to treat tumors or prevent them from spreading, is in actuality not a usable method of treatment, since it can cause premature aging.[26] However, restoring endogenous p53 function holds a lot of promise.[27] Loss of p53 creates genomic instability that most often results in the aneuploidy phenotype.[28]
Certain pathogens can also affect the p53 protein that the TP53 gene expresses. One such example, human papillomavirus (HPV), encodes a protein, E6, which binds the p53 protein and inactivates it. This, in synergy with the inactivation of another cell cycle regulator, pRb, by the HPV protein E7, allows for repeated cell division manifested in the clinical disease of warts. Certain HPV types, in particular types 16 and 18, can also lead to progression from a benign wart to low or high-grade cervical dysplasia, which are reversible forms of precancerous lesions. Persistent infection of the cervix over the years can cause irreversible changes leading to carcinoma in situ and eventually invasive cervical cancer. This results from the effects of HPV genes, particularly those encoding E6 and E7, which are the two viral oncoproteins that are preferentially retained and expressed in cervical cancers by integration of the viral DNA into the host genome.[29]
In healthy humans, the p53 protein is continually produced and degraded in the cell. The degradation of the p53 protein is, as mentioned, associated with MDM2 binding. In a negative feedback loop, MDM2 is itself induced by the p53 protein. However, mutant p53 proteins often do not induce MDM2, and are thus able to accumulate at very high concentrations. Worse, mutant p53 protein itself can inhibit normal p53 protein levels.
Discovery
p53 was identified in 1979 by Lionel Crawford, David P. Lane, Arnold Levine, and Lloyd Old, working at Imperial Cancer Research Fund (UK) Princeton University/UMDNJ (Cancer Institute of New Jersey), and Sloan-Kettering Memorial Hospital, respectively. It had been hypothesized to exist before as the target of the SV40 virus, a strain that induced development of tumors. The TP53 gene from the mouse was first cloned by Peter Chumakov of the Russian Academy of Sciences in 1982,[30] and independently in 1983 by Moshe Oren in collaboration with David Givol (Weizmann Institute of Science).[31][32] The human TP53 gene was cloned in 1984[1] and the full length clone in 1985.[33]
It was initially presumed to be an oncogene due to the use of mutated cDNA following purification of tumour cell mRNA. Its character as a tumor suppressor gene was finally revealed in 1989 by Bert Vogelstein working at Johns Hopkins School of Medicine.[34]
Warren Maltzman, of the Waksman Institute of Rutgers University first demonstrated that TP53 was responsive to DNA damage in the form of ultraviolet radiation.[35] In a series of publications in 1991-92, Michael Kastan, Johns Hopkins University, reported that TP53 was a critical part of a signal transduction pathway that helped cells respond to DNA damage.[36]
In 1992, Wafik El-Deiry when he was working with Bert Vogelstein at Johns Hopkins University identified the consensus sequence, to which human p53 could bind, by immunoprecipitating human genomic DNA that could be bound by baculovirus-produced human p53 protein. This sequence was published in the first issue of the journal Nature Genetics in 1992 in work that is highly cited. The consensus sequence is 5'-RRRCWWGYYY-N(0-13)-RRRCWWGYYY-3' and is located in the regulatory regions of genes that are activated by the p53 transcription factor. The presence of p53 response elements in or around genes (promoters, upstream sequences, introns) is a powerful predictor of regulation and activation of a particular gene by p53.
In 1993, p53 was voted molecule of the year by Science magazine.[37]
That same year, 1993, Wafik El-Deiry when he was working with Bert Vogelstein at Johns Hopkins University discovered p21(WAF1) as a gene regulated directly by p53. This work was reported in the most highly cited paper ever published in the journal Cell, and provided a molecular mechanism by which mammalian cells undergo growth arrest when damaged. The p21(WAF1) protein binds directly to cyclin-CDK complexes that drive forward the cell cycle and inhibits their kinase activity thereby causing cell cycle arrest to allow repair to take place. p21 can also mediate growth arrest associated with differentiation and a more permanent growth arrest associated with cellular senescence. The p21 gene contains several p53 response elements that mediate direct binding of the p53 protein, resulting in transcriptional activation of the gene encoding the p21(WAF1) protein.
Interactions
p53 has been shown to interact with
- ANKRD2[38]
- Aprataxin[39]
- Ataxia telangiectasia and Rad3 related[40][41]
- Ataxia telangiectasia mutated[40]
- ATF3,[42]
- Aurora A kinase[43]
- BAK1[44]
- BARD1[45]
- Bloom syndrome protein[46]
- BRCA1[45]
- BRCA2[45]
- BRCC3[45]
- BRE[45]
- CCAAT/enhancer binding protein zeta[47]
- CDC14A[48]
- Cdk1[49]
- CFLAR[50]
- CHEK1[46]
- CREB-binding protein[51]
- CREB1[52]
- Cyclin H[53]
- Cyclin-dependent kinase 7[53][54]
- DNA-PKcs[41]
- E4F1[55]
- EFEMP2[56]
- EP300[57]
- ERCC6[58]
- GNL3[59]
- GPS2[60]
- GSK3B[61]
- Heat shock protein 90kDa alpha (cytosolic), member A1[62]
- HIF1A[63]
- HIPK1[64]
- HIPK2[65]
- HMGB1[66]
- HSPA9[67]
- Huntingtin[68]
- ING1[69]
- ING4[70][71]
- ING5[70] ELL,[72]
- IκBα[73]
- KPNB1[62]
- Mdm2[51]
- MDM4[74]
- MED1[75]
- Mitogen-activated protein kinase 9[76]
- MNAT1[54]
- Multisynthetase complex auxiliary component p38[77]
- NDN[78]
- Nucleolin[79]
- NUMB[80]
- P16[55]
- PARC[81]
- PARP1[39]
- PIAS1[56] CDC14B[48]
- PIN1[82]
- PLAGL1[83]
- PLK3[84]
- PRKRA[85]
- Prohibitin[86]
- Promyelocytic leukemia protein[87]
- Protein kinase R[88]
- PSME3[89]
- PTEN[90]
- PTK2[91]
- PTTG1[92]
- RAD51[45]
- RCHY1[93]
- Replication protein A1[94]
- RPL11[95]
- S100B,[96]
- Small ubiquitin-related modifier 1[97]
- SMARCA4,[98]
- SMARCB1[98]
- SMN1[99]
- TATA binding protein[100]
- TFAP2A[101]
- TFDP1[102]
- TOP1[103]
- TOP2A[104]
- TP53BP1[46]
- TP53BP2,[105] TOP2B,[104]
- TP53INP1[106]
- TSG101[107]
- UBE2A[108]
- UBE2I[56]
- Ubiquitin C[77]
- USP7[109]
- Werner syndrome ATP-dependent helicase[110]
- WWOX[111]
- XPB[58]
- Y box binding protein 1[38]
- YPEL3[112]
- YWHAZ[113]
- Zif268[114]
- ZNF148[115]
References
- ^ a b c Matlashewski G, Lamb P, Pim D, Peacock J, Crawford L, Benchimol S (December 1984). "Isolation and characterization of a human p53 cDNA clone: expression of the human p53 gene". EMBO J. 3 (13): 3257–62. PMC 557846. PMID 6396087. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=557846.
- ^ a b Isobe M, Emanuel BS, Givol D, Oren M, Croce CM (1986). "Localization of gene for human p53 tumour antigen to band 17p13". Nature 320 (6057): 84–5. doi:10.1038/320084a0. PMID 3456488.
- ^ a b Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B (June 1991). "Identification of p53 as a sequence-specific DNA-binding protein". Science 252 (5013): 1708–11. doi:10.1126/science.2047879. PMID 2047879.
- ^ a b McBride OW, Merry D, Givol D (1986). "The gene for human p53 cellular tumor antigen is located on chromosome 17 short arm (17p13)". Proc. Natl. Acad. Sci. U.S.A. 83 (1): 130–134. doi:10.1073/pnas.83.1.130. PMC 322805. PMID 3001719. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=322805.
- ^ Read, A. P.; Strachan, T. (1999). "Chapter 18: Cancer Genetics". Human molecular genetics 2. New York: Wiley.
- ^ Ziemer MA, Mason A, Carlson DM (September 1982). "Cell-free translations of proline-rich protein mRNAs". J. Biol. Chem. 257 (18): 11176–80. PMID 7107651. http://www.jbc.org/content/257/18/11176.abstract.
- ^ May, P. and May, E. (1999). Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene, 18, 7621–36.
- ^ "OrthoMaM phylogenetic marker: TP53 coding sequence". http://www.orthomam.univ-montp2.fr/orthomam/data/cds/detailMarkers/ENSG00000141510_TP53.xml.
- ^ Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, Brenna SM, Ciotti M, Das BR, Del Mistro A, Dybikowska A, Giuliano AR, Gudleviciene Z, Gyllensten U, Haws AL, Helland A, Herrington CS, Hildesheim A, Humbey O, Jee SH, Kim JW, Madeleine MM, Menczer J, Ngan HY, Nishikawa A, Niwa Y, Pegoraro R, Pillai MR, Ranzani G, Rezza G, Rosenthal AN, Roychoudhury S, Saranath D, Schmitt VM, Sengupta S, Settheetham-Ishida W, Shirasawa H, Snijders PJ, Stoler MH, Suárez-Rincón AE, Szarka K, Tachezy R, Ueda M, van der Zee AG, von Knebel Doeberitz M, Wu MT, Yamashita T, Zehbe I, Blettner M. (2009). "TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies.". Lancet Oncology 10 (2): 772–784. doi:10.1016/S1470-2045(09)70187-1. PMID 19625214.
- ^ Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L (August 1998). "The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression". EMBO J. 17 (16): 4668–79. doi:10.1093/emboj/17.16.4668. PMC 1170796. PMID 9707426. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1170796.
- ^ a b Larsen S, Yokochi T, Isogai E, Nakamura Y, Ozaki T, Nakagawara A (February 2010). "LMO3 interacts with p53 and inhibits its transcriptional activity". Biochem. Biophys. Res. Commun. 392 (3): 252–7. doi:10.1016/j.bbrc.2009.12.010. PMID 19995558. http://www.ncbi.nlm.nih.gov/pubmed/19995558.
- ^ Harms KL, Chen X (2005). "The C terminus of p53 family proteins is a cell fate determinant". Mol. Cell. Biol. 25 (5): 2014–30. doi:10.1128/MCB.25.5.2014-2030.2005. PMC 549381. PMID 15713654. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=549381.
- ^ Piskacek S, Gregor M, Nemethova M, Grabner M, Kovarik P, Piskacek M (June 2007). "Nine-amino-acid transactivation domain: establishment and prediction utilities". Genomics 89 (6): 756–68. doi:10.1016/j.ygeno.2007.02.003. PMID 17467953.
- ^ Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL (August 1997). "Induced alpha helix in the VP16 activation domain upon binding to a human TAF". Science 277 (5330): 1310–3. doi:10.1126/science.277.5330.1310. PMID 9271577.; Uesugi M, Verdine GL (December 1999). "The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2". Proc. Natl. Acad. Sci. U.S.A. 96 (26): 14801–6. doi:10.1073/pnas.96.26.14801. PMC 24728. PMID 10611293. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=24728.; Choi Y, Asada S, Uesugi M (May 2000). "Divergent hTAFII31-binding motifs hidden in activation domains". J. Biol. Chem. 275 (21): 15912–6. doi:10.1074/jbc.275.21.15912. PMID 10821850.; Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L (April 1999). "Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains". Oncogene 18 (14): 2405–10. doi:10.1038/sj.onc.1202539. PMID 10327062.; Lin J, Chen J, Elenbaas B, Levine AJ (May 1994). "Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein". Genes Dev. 8 (10): 1235–46. doi:10.1101/gad.8.10.1235. PMID 7926727.
- ^ Piskacek S, Gregor M, Nemethova M, Grabner M, Kovarik P, Piskacek M (June 2007). "Nine-amino-acid transactivation domain: establishment and prediction utilities". Genomics 89 (6): 756–68. doi:10.1016/j.ygeno.2007.02.003. PMID 17467953.; Piskacek M (2009-11-05). "9aaTAD is a common transactivation domain recruits multiple general coactivators TAF9, MED15, CBP/p300 and GCN5". Nature Precedings Pre-publication. doi:10.1038/npre.2009.3488.2.; Piskacek M (2009-11-05). "9aaTADs mimic DNA to interact with a pseudo-DNA Binding Domain KIX of Med15 (Molecular Chameleons)". Nature Precedings Pre-publication. doi:10.1038/npre.2009.3939.1.; Piskacek M; Piskacek, Martin (2009-11-20). "9aaTAD Prediction result (2006)". Nature Precedings Pre-publication. doi:10.1038/npre.2009.3984.1.
- ^ The prediction for 9aaTADs (for both acidic and hydrophilic transactivation domains) is available online from ExPASy http://us.expasy.org/tools/ and EMBnet Spain http://www.es.embnet.org/Services/EMBnetAT/htdoc/9aatad/
- ^ Bell S, Klein C, Müller L, Hansen S, Buchner J (2002). "p53 contains large unstructured regions in its native state". J. Mol. Biol. 322 (5): 917–27. doi:10.1016/S0022-2836(02)00848-3. PMID 12367518.
- ^ National Center for Biotechnology Information. "The p53 tumor suppressor protein". Genes and Disease. United States National Institutes of Health. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View..ShowSection&rid=gnd.section.107. Retrieved 2008-05-28.
- ^ Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH (1998). "p14ARF links the tumour suppressors RB and p53". Nature 395 (6698): 124–5. doi:10.1038/25867. PMID 9744267.
- ^ Hu W, Feng Z, Teresky AK, Levine AJ (November 2007). "p53 regulates maternal reproduction through LIF". Nature 450 (7170): 721–4. doi:10.1038/nature05993. PMID 18046411.
- ^ "Genome's guardian gets a tan started". New Scientist. March 17, 2007. http://www.newscientist.com/channel/health/mg19325955.800-genomes-guardian-gets-a-tan-started.html. Retrieved 2007-03-29.
- ^ Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter SR, Fisher DE (2007). "Central role of p53 in the suntan response and pathologic hyperpigmentation". Cell 128 (5): 853–64. doi:10.1016/j.cell.2006.12.045. PMID 17350573.
- ^ Han ES, Muller FL, Pérez VI, Qi W, Liang H, Xi L, Fu C, Doyle E, Hickey M, Cornell J, Epstein CJ, Roberts LJ, Van Remmen H, Richardson A (June 2008). "The in vivo gene expression signature of oxidative stress". Physiol. Genomics 34 (1): 112–26. doi:10.1152/physiolgenomics.00239.2007. PMC 2532791. PMID 18445702. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2532791.
- ^ Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober (March 2008). "Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice". Circ. Res. 102 (6): 703–10. doi:10.1161/CIRCRESAHA.107.164558. PMID 18239138.
- ^ Hollstein M, Sidransky D, Vogelstein B, Harris CC (1991). "p53 mutations in human cancers". Science 253 (5015): 49–53. doi:10.1126/science.1905840. PMID 1905840.
- ^ Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee Park S, Thompson T, Karsenty G, Bradley A, Donehower LA (2002). "p53 mutant mice that display early ageing-associated phenotypes". Nature 415 (6867): 45–53. doi:10.1038/415045a. PMID 11780111.
- ^ Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T (2007). "Restoration of p53 function leads to tumour regression in vivo". Nature 445 (7128): 661–5. doi:10.1038/nature05541. PMID 17251932.
- ^ Clemens A. Schmitt; Fridman, JS; Yang, M; Baranov, E; Hoffman, RM; Lowe, SW (April 2002). "Dissecting p53 tumor suppressor functions in vivo". Cancer Cell 1 (3): 289–298. doi:10.1016/S1535-6108(02)00047-8. PMID 12086865.
- ^ Angeletti PC, Zhang L, Wood C (2008). "The viral etiology of AIDS-associated malignancies". Adv. Pharmacol. 56: 509–57. doi:10.1016/S1054-3589(07)56016-3. PMC 2149907. PMID 18086422. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2149907.
- ^ Chumakov PM, Iotsova VS, Georgiev GP (1982). "[Isolation of a plasmid clone containing the mRNA sequence for mouse nonviral T-antigen]" (in Russian). Dokl. Akad. Nauk SSSR 267 (5): 1272–5. PMID 6295732.
- ^ Oren M, Levine AJ (January 1983). "Molecular cloning of a cDNA specific for the murine p53 cellular tumor antigen". Proc. Natl. Acad. Sci. U.S.A. 80 (1): 56–9. doi:10.1073/pnas.80.1.56. PMC 393308. PMID 6296874. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=393308.
- ^ Zakut-Houri R, Oren M, Bienz B, Lavie V, Hazum S, Givol D (1983). "A single gene and a pseudogene for the cellular tumour antigen p53". Nature 306 (5943): 594–7. doi:10.1038/306594a0. PMID 6646235.
- ^ Zakut-Houri R, Bienz-Tadmor B, Givol D, Oren M (May 1985). "Human p53 cellular tumor antigen: cDNA sequence and expression in COS cells". EMBO J. 4 (5): 1251–5. PMC 554332. PMID 4006916. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=554332.
- ^ Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B (April 1989). "Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas". Science 244 (4901): 217–21. doi:10.1126/science.264998. PMID 2649981.
- ^ Maltzman W, Czyzyk L (September 1984). "UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells". Mol. Cell. Biol. 4 (9): 1689–94. PMC 368974. PMID 6092932. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=368974.
- ^ Kastan MB, Kuerbitz SJ (December 1993). "Control of G1 arrest after DNA damage". Environ. Health Perspect. 101 Suppl 5: 55–8. doi:10.2307/3431842. JSTOR 3431842. PMC 1519427. PMID 8013425. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1519427.
- ^ Koshland DE (December 1993). "Molecule of the year". Science 262 (5142): 1953. doi:10.1126/science.8266084. PMID 8266084.
- ^ a b Kojic, Snezana; Medeot Elisa, Guccione Ernesto, Krmac Helena, Zara Ivano, Martinelli Valentina, Valle Giorgio, Faulkner Georgine (May. 2004). "The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle". J. Mol. Biol. 339 (2): 313–25. doi:10.1016/j.jmb.2004.03.071. PMID 15136035.
- ^ a b Gueven, Nuri; Becherel Olivier J, Kijas Amanda W, Chen Philip, Howe Orla, Rudolph Jeanette H, Gatti Richard, Date Hidetoshi, Onodera Osamu, Taucher-Scholz Gisela, Lavin Martin F (May. 2004). "Aprataxin, a novel protein that protects against genotoxic stress". Hum. Mol. Genet. 13 (10): 1081–93. doi:10.1093/hmg/ddh122. PMID 15044383.
- ^ a b Fabbro, Megan; Savage Kienan, Hobson Karen, Deans Andrew J, Powell Simon N, McArthur Grant A, Khanna Kum Kum (Jul. 2004). "BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage". J. Biol. Chem. 279 (30): 31251–8. doi:10.1074/jbc.M405372200. PMID 15159397.
- ^ a b Kim, S T; Lim D S, Canman C E, Kastan M B (Dec. 1999). "Substrate specificities and identification of putative substrates of ATM kinase family members". J. Biol. Chem. 274 (53): 37538–43. doi:10.1074/jbc.274.53.37538. PMID 10608806.
- ^ Stelzl, Ulrich; Worm Uwe, Lalowski Maciej, Haenig Christian, Brembeck Felix H, Goehler Heike, Stroedicke Martin, Zenkner Martina, Schoenherr Anke, Koeppen Susanne, Timm Jan, Mintzlaff Sascha, Abraham Claudia, Bock Nicole, Kietzmann Silvia, Goedde Astrid, Toksöz Engin, Droege Anja, Krobitsch Sylvia, Korn Bernhard, Birchmeier Walter, Lehrach Hans, Wanker Erich E (Sep. 2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. PMID 16169070.
- ^ Chen, Shih-Shun; Chang Pi-Chu, Cheng Yu-Wen, Tang Fen-Mei, Lin Young-Sun (Sep. 2002). "Suppression of the STK15 oncogenic activity requires a transactivation-independent p53 function". EMBO J. 21 (17): 4491–9. doi:10.1093/emboj/cdf409. PMC 126178. PMID 12198151. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=126178.
- ^ Leu, J I-Ju; Dumont Patrick, Hafey Michael, Murphy Maureen E, George Donna L (May. 2004). "Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex". Nat. Cell Biol. 6 (5): 443–50. doi:10.1038/ncb1123. PMID 15077116.
- ^ a b c d e f Dong, Yuanshu; Hakimi Mohamed-Ali, Chen Xiaowei, Kumaraswamy Easwari, Cooch Neil S, Godwin Andrew K, Shiekhattar Ramin (Nov. 2003). "Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair". Mol. Cell 12 (5): 1087–99. doi:10.1016/S1097-2765(03)00424-6. PMID 14636569.
- ^ a b c Sengupta, Sagar; Robles Ana I, Linke Steven P, Sinogeeva Natasha I, Zhang Ran, Pedeux Remy, Ward Irene M, Celeste Arkady, Nussenzweig André, Chen Junjie, Halazonetis Thanos D, Harris Curtis C (Sep. 2004). "Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest". J. Cell Biol. 166 (6): 801–13. doi:10.1083/jcb.200405128. PMC 2172115. PMID 15364958. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2172115.
- ^ Uramoto, Hidetaka; Izumi Hiroto, Nagatani Gunji, Ohmori Haruki, Nagasue Naofumi, Ise Tomoko, Yoshida Takeshi, Yasumoto Kosei, Kohno Kimitoshi (Apr. 2003). "Physical interaction of tumour suppressor p53/p73 with CCAAT-binding transcription factor 2 (CTF2) and differential regulation of human high-mobility group 1 (HMG1) gene expression". Biochem. J. 371 (Pt 2): 301–10. doi:10.1042/BJ20021646. PMC 1223307. PMID 12534345. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1223307.
- ^ a b Li, L; Ljungman M, Dixon J E (Jan. 2000). "The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53". J. Biol. Chem. 275 (4): 2410–4. doi:10.1074/jbc.275.4.2410. PMID 10644693.
- ^ Luciani, M G; Hutchins J R, Zheleva D, Hupp T R (Jul. 2000). "The C-terminal regulatory domain of p53 contains a functional docking site for cyclin A". J. Mol. Biol. 300 (3): 503–18. doi:10.1006/jmbi.2000.3830. PMID 10884347.
- ^ Abedini, Mohammad R; Muller Emilie J, Brun Jan, Bergeron Richard, Gray Douglas A, Tsang Benjamin K (Jun. 2008). "Cisplatin induces p53-dependent FLICE-like inhibitory protein ubiquitination in ovarian cancer cells". Cancer Res. 68 (12): 4511–7. doi:10.1158/0008-5472.CAN-08-0673. PMID 18559494.
- ^ a b Ito, Akihiro; Kawaguchi Yoshiharu, Lai Chun-Hsiang, Kovacs Jeffrey J, Higashimoto Yuichiro, Appella Ettore, Yao Tso-Pang (Nov. 2002). "MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation". EMBO J. 21 (22): 6236–45. doi:10.1093/emboj/cdf616. PMC 137207. PMID 12426395. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=137207.
- ^ Giebler, H A; Lemasson I, Nyborg J K (Jul. 2000). "p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation". Mol. Cell. Biol. 20 (13): 4849–58. doi:10.1128/MCB.20.13.4849-4858.2000. PMC 85936. PMID 10848610. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=85936.
- ^ a b Schneider, E; Montenarh M, Wagner P (Nov. 1998). "Regulation of CAK kinase activity by p53". Oncogene 17 (21): 2733–41. doi:10.1038/sj.onc.1202504. PMID 9840937.
- ^ a b Ko, L J; Shieh S Y, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan Z Q (Dec. 1997). "p53 is phosphorylated by CDK7-cyclin H in a p36MAT1-dependent manner". Mol. Cell. Biol. 17 (12): 7220–9. PMC 232579. PMID 9372954. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=232579.
- ^ a b Rizos, Helen; Diefenbach Eve, Badhwar Prerna, Woodruff Sarah, Becker Therese M, Rooney Robert J, Kefford Richard F (Feb. 2003). "Association of p14ARF with the p120E4F transcriptional repressor enhances cell cycle inhibition". J. Biol. Chem. 278 (7): 4981–9. doi:10.1074/jbc.M210978200. PMID 12446718.
- ^ a b c Gallagher, W M; Argentini M, Sierra V, Bracco L, Debussche L, Conseiller E (Jun. 1999). "MBP1: a novel mutant p53-specific protein partner with oncogenic properties". Oncogene 18 (24): 3608–16. doi:10.1038/sj.onc.1202937. PMID 10380882.
- ^ Livengood, Jill A; Scoggin Kirsten E S, Van Orden Karen, McBryant Steven J, Edayathumangalam Rajeswari S, Laybourn Paul J, Nyborg Jennifer K (Mar. 2002). "p53 Transcriptional activity is mediated through the SRC1-interacting domain of CBP/p300". J. Biol. Chem. 277 (11): 9054–61. doi:10.1074/jbc.M108870200. PMID 11782467.
- ^ a b Wang, X W; Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G (Jun. 1995). "p53 modulation of TFIIH-associated nucleotide excision repair activity". Nat. Genet. 10 (2): 188–95. doi:10.1038/ng0695-188. PMID 7663514.
- ^ Tsai, Robert Y L; McKay Ronald D G (Dec. 2002). "A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells". Genes Dev. 16 (23): 2991–3003. doi:10.1101/gad.55671. PMC 187487. PMID 12464630. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=187487.
- ^ Peng, Y C; Kuo F, Breiding D E, Wang Y F, Mansur C P, Androphy E J (Sep. 2001). "AMF1 (GPS2) modulates p53 transactivation". Mol. Cell. Biol. 21 (17): 5913–24. doi:10.1128/MCB.21.17.5913-5924.2001. PMC 87310. PMID 11486030. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=87310.
- ^ Watcharasit, Piyajit; Bijur Gautam N, Zmijewski Jaroslaw W, Song Ling, Zmijewska Anna, Chen Xinbin, Johnson Gail V W, Jope Richard S (Jun. 2002). "Direct, activating interaction between glycogen synthase kinase-3beta and p53 after DNA damage". Proc. Natl. Acad. Sci. U.S.A. 99 (12): 7951–5. doi:10.1073/pnas.122062299. PMC 123001. PMID 12048243. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=123001.
- ^ a b Akakura, S; Yoshida M, Yoneda Y, Horinouchi S (May. 2001). "A role for Hsc70 in regulating nucleocytoplasmic transport of a temperature-sensitive p53 (p53Val-135)". J. Biol. Chem. 276 (18): 14649–57. doi:10.1074/jbc.M100200200. PMID 11297531.
- ^ Chen, Delin; Li Muyang, Luo Jianyuan, Gu Wei (Apr. 2003). "Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function". J. Biol. Chem. 278 (16): 13595–8. doi:10.1074/jbc.C200694200. PMID 12606552.
- ^ Kondo, Seiji; Lu Ying, Debbas Michael, Lin Athena W, Sarosi Ildiko, Itie Annick, Wakeham Andrew, Tuan JoAnn, Saris Chris, Elliott Gary, Ma Weili, Benchimol Samuel, Lowe Scott W, Mak Tak Wah, Thukral Sushil K (Apr. 2003). "Characterization of cells and gene-targeted mice deficient for the p53-binding kinase homeodomain-interacting protein kinase 1 (HIPK1)". Proc. Natl. Acad. Sci. U.S.A. 100 (9): 5431–6. doi:10.1073/pnas.0530308100. PMC 154362. PMID 12702766. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=154362.
- ^ Hofmann, Thomas G; Möller Andreas, Sirma Hüaeyin, Zentgraf Hanswalter, Taya Yoichi, Dröge Wulf, Will Hans, Schmitz M Lienhard (Jan. 2002). "Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2". Nat. Cell Biol. 4 (1): 1–10. doi:10.1038/ncb715. PMID 11740489.
- ^ Imamura, T; Izumi H, Nagatani G, Ise T, Nomoto M, Iwamoto Y, Kohno K (Mar. 2001). "Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein". J. Biol. Chem. 276 (10): 7534–40. doi:10.1074/jbc.M008143200. PMID 11106654.
- ^ Wadhwa, Renu; Yaguchi Tomoko, Hasan Md K, Mitsui Youji, Reddel Roger R, Kaul Sunil C (Apr. 2002). "Hsp70 family member, mot-2/mthsp70/GRP75, binds to the cytoplasmic sequestration domain of the p53 protein". Exp. Cell Res. 274 (2): 246–53. doi:10.1006/excr.2002.5468. PMID 11900485.
- ^ Steffan, J S; Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M (Jun. 2000). "The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription". Proc. Natl. Acad. Sci. U.S.A. 97 (12): 6763–8. doi:10.1073/pnas.100110097. PMC 18731. PMID 10823891. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=18731.
- ^ Leung, Ka Man; Po Lai See, Tsang Fan Cheung, Siu Wai Yi, Lau Anita, Ho Horace T B, Poon Randy Y C (Sep. 2002). "The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2". Cancer Res. 62 (17): 4890–3. PMID 12208736.
- ^ a b Shiseki, Masayuki; Nagashima Makoto, Pedeux Remy M, Kitahama-Shiseki Mariko, Miura Koh, Okamura Shu, Onogi Hitoshi, Higashimoto Yuichiro, Appella Ettore, Yokota Jun, Harris Curtis C (May. 2003). "p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity". Cancer Res. 63 (10): 2373–8. PMID 12750254.
- ^ Tsai, Kuo-Wang; Tseng Hsiao-Chun, Lin Wen-Chang (Oct. 2008). "Two wobble-splicing events affect ING4 protein subnuclear localization and degradation". Exp. Cell Res. 314 (17): 3130–41. doi:10.1016/j.yexcr.2008.08.002. PMID 18775696.
- ^ Shinobu, N; Maeda T, Aso T, Ito T, Kondo T, Koike K, Hatakeyama M (Jun. 1999). "Physical interaction and functional antagonism between the RNA polymerase II elongation factor ELL and p53". J. Biol. Chem. 274 (24): 17003–10. doi:10.1074/jbc.274.24.17003. PMID 10358050.
- ^ Chang, Nan-Shan (Mar. 2002). "The non-ankyrin C terminus of Ikappa Balpha physically interacts with p53 in vivo and dissociates in response to apoptotic stress, hypoxia, DNA damage, and transforming growth factor-beta 1-mediated growth suppression". J. Biol. Chem. 277 (12): 10323–31. doi:10.1074/jbc.M106607200. PMID 11799106.
- ^ Badciong, James C; Haas Arthur L (Dec. 2002). "MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination". J. Biol. Chem. 277 (51): 49668–75. doi:10.1074/jbc.M208593200. PMID 12393902.
- ^ Frade, R; Balbo M, Barel M (Dec. 2000). "RB18A, whose gene is localized on chromosome 17q12-q21.1, regulates in vivo p53 transactivating activity". Cancer Res. 60 (23): 6585–9. PMID 11118038.
- ^ Hu, M C; Qiu W R, Wang Y P (Nov. 1997). "JNK1, JNK2 and JNK3 are p53 N-terminal serine 34 kinases". Oncogene 15 (19): 2277–87. doi:10.1038/sj.onc.1201401. PMID 9393873.
- ^ a b Han, Jung Min; Park Bum-Joon, Park Sang Gyu, Oh Young Sun, Choi So Jung, Lee Sang Won, Hwang Soon-Kyung, Chang Seung-Hee, Cho Myung-Haing, Kim Sunghoon (Aug. 2008). "AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53". Proc. Natl. Acad. Sci. U.S.A. 105 (32): 11206–11. doi:10.1073/pnas.0800297105. PMC 2516205. PMID 18695251. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2516205.
- ^ Taniura, H; Matsumoto K, Yoshikawa K (Jun. 1999). "Physical and functional interactions of neuronal growth suppressor necdin with p53". J. Biol. Chem. 274 (23): 16242–8. doi:10.1074/jbc.274.23.16242. PMID 10347180.
- ^ Daniely, Yaron; Dimitrova Diana D, Borowiec James A (Aug. 2002). "Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation". Mol. Cell. Biol. 22 (16): 6014–22. doi:10.1128/MCB.22.16.6014-6022.2002. PMC 133981. PMID 12138209. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=133981.
- ^ Colaluca, Ivan N; Tosoni Daniela, Nuciforo Paolo, Senic-Matuglia Francesca, Galimberti Viviana, Viale Giuseppe, Pece Salvatore, Di Fiore Pier Paolo (Jan. 2008). "NUMB controls p53 tumour suppressor activity". Nature 451 (7174): 76–80. doi:10.1038/nature06412. PMID 18172499.
- ^ Nikolaev, Anatoly Y; Li Muyang, Puskas Norbert, Qin Jun, Gu Wei (Jan. 2003). "Parc: a cytoplasmic anchor for p53". Cell 112 (1): 29–40. doi:10.1016/S0092-8674(02)01255-2. PMID 12526791.
- ^ Wulf, Gerburg M; Liou Yih-Cherng, Ryo Akihide, Lee Sam W, Lu Kun Ping (Dec. 2002). "Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage". J. Biol. Chem. 277 (50): 47976–9. doi:10.1074/jbc.C200538200. PMID 12388558.
- ^ Huang, S M; Schönthal A H, Stallcup M R (Apr. 2001). "Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1". Oncogene 20 (17): 2134–43. doi:10.1038/sj.onc.1204298. PMID 11360197.
- ^ Xie, S; Wu H, Wang Q, Cogswell J P, Husain I, Conn C, Stambrook P, Jhanwar-Uniyal M, Dai W (Nov. 2001). "Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway". J. Biol. Chem. 276 (46): 43305–12. doi:10.1074/jbc.M106050200. PMID 11551930.
- ^ Simons, A; Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V (Jan. 1997). "PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb". Oncogene 14 (2): 145–55. doi:10.1038/sj.onc.1200825. PMID 9010216.
- ^ Fusaro, Gina; Dasgupta Piyali, Rastogi Shipra, Joshi Bharat, Chellappan Srikumar (Nov. 2003). "Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling". J. Biol. Chem. 278 (48): 47853–61. doi:10.1074/jbc.M305171200. PMID 14500729.
- ^ Kurki, Sari; Latonen Leena, Laiho Marikki (Oct. 2003). "Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization". J. Cell. Sci. 116 (Pt 19): 3917–25. doi:10.1242/jcs.00714. PMID 12915590.
- ^ Cuddihy, A R; Wong A H, Tam N W, Li S, Koromilas A E (Apr. 1999). "The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro". Oncogene 18 (17): 2690–702. doi:10.1038/sj.onc.1202620. PMID 10348343.
- ^ Zhang, Zhuo; Zhang Ruiwen (Mar. 2008). "Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation". EMBO J. 27 (6): 852–64. doi:10.1038/emboj.2008.25. PMC 2265109. PMID 18309296. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2265109.
- ^ Freeman, Daniel J; Li Andrew G, Wei Gang, Li Heng-Hong, Kertesz Nathalie, Lesche Ralf, Whale Andrew D, Martinez-Diaz Hilda, Rozengurt Nora, Cardiff Robert D, Liu Xuan, Wu Hong (Feb. 2003). "PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms". Cancer Cell 3 (2): 117–30. doi:10.1016/S1535-6108(03)00021-7. PMID 12620407.
- ^ Lim, Ssang-Taek; Chen Xiao Lei, Lim Yangmi, Hanson Dan A, Vo Thanh-Trang, Howerton Kyle, Larocque Nicholas, Fisher Susan J, Schlaepfer David D, Ilic Dusko (Jan. 2008). "Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation". Mol. Cell 29 (1): 9–22. doi:10.1016/j.molcel.2007.11.031. PMC 2234035. PMID 18206965. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2234035.
- ^ Bernal, Juan A; Luna Rosa, Espina Agueda, Lázaro Icíar, Ramos-Morales Francisco, Romero Francisco, Arias Carmen, Silva Augusto, Tortolero María, Pintor-Toro José A (Oct. 2002). "Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis". Nat. Genet. 32 (2): 306–11. doi:10.1038/ng997. PMID 12355087.
- ^ Leng, Roger P; Lin Yunping, Ma Weili, Wu Hong, Lemmers Benedicte, Chung Stephen, Parant John M, Lozano Guillermina, Hakem Razqallah, Benchimol Samuel (Mar. 2003). "Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation". Cell 112 (6): 779–91. doi:10.1016/S0092-8674(03)00193-4. PMID 12654245.
- ^ Romanova, Larisa Y; Willers Henning, Blagosklonny Mikhail V, Powell Simon N (Dec. 2004). "The interaction of p53 with replication protein A mediates suppression of homologous recombination". Oncogene 23 (56): 9025–33. doi:10.1038/sj.onc.1207982. PMID 15489903.
- ^ Zhang, Yanping; Wolf Gabrielle White, Bhat Krishna, Jin Aiwen, Allio Theresa, Burkhart William A, Xiong Yue (Dec. 2003). "Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway". Mol. Cell. Biol. 23 (23): 8902–12. doi:10.1128/MCB.23.23.8902-8912.2003. PMC 262682. PMID 14612427. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=262682.
- ^ Lin, Jing; Yang Qingyuan, Yan Zhe, Markowitz Joseph, Wilder Paul T, Carrier France, Weber David J (Aug. 2004). "Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells". J. Biol. Chem. 279 (32): 34071–7. doi:10.1074/jbc.M405419200. PMID 15178678.
- ^ Minty, A; Dumont X, Kaghad M, Caput D (Nov. 2000). "Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif". J. Biol. Chem. 275 (46): 36316–23. doi:10.1074/jbc.M004293200. PMID 10961991.
- ^ a b Lee, Daeyoup; Kim Jin Woo, Seo Taegun, Hwang Sun Gwan, Choi Eui-Ju, Choe Joonho (Jun. 2002). "SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription". J. Biol. Chem. 277 (25): 22330–7. doi:10.1074/jbc.M111987200. PMID 11950834.
- ^ Young, Philip J; Day Patricia M, Zhou Jianhua, Androphy Elliot J, Morris Glenn E, Lorson Christian L (Jan. 2002). "A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy". J. Biol. Chem. 277 (4): 2852–9. doi:10.1074/jbc.M108769200. PMID 11704667.
- ^ Seto, E; Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T (Dec. 1992). "Wild-type p53 binds to the TATA-binding protein and represses transcription". Proc. Natl. Acad. Sci. U.S.A. 89 (24): 12028–32. doi:10.1073/pnas.89.24.12028. PMC 50691. PMID 1465435. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=50691.
- ^ McPherson, Lisa A; Loktev Alexander V, Weigel Ronald J (Nov. 2002). "Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53". J. Biol. Chem. 277 (47): 45028–33. doi:10.1074/jbc.M208924200. PMID 12226108.
- ^ Sørensen, T S; Girling R, Lee C W, Gannon J, Bandara L R, La Thangue N B (Oct. 1996). "Functional interaction between DP-1 and p53". Mol. Cell. Biol. 16 (10): 5888–95. PMC 231590. PMID 8816502. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=231590.
- ^ Gobert, C; Skladanowski A, Larsen A K (Aug. 1999). "The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53". Proc. Natl. Acad. Sci. U.S.A. 96 (18): 10355–60. doi:10.1073/pnas.96.18.10355. PMC 17892. PMID 10468612. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=17892.
- ^ a b Cowell, I G; Okorokov A L, Cutts S A, Padget K, Bell M, Milner J, Austin C A (Feb. 2000). "Human topoisomerase IIalpha and IIbeta interact with the C-terminal region of p53". Exp. Cell Res. 255 (1): 86–94. doi:10.1006/excr.1999.4772. PMID 10666337.
- ^ Iwabuchi, K; Bartel P L, Li B, Marraccino R, Fields S (Jun. 1994). "Two cellular proteins that bind to wild-type but not mutant p53". Proc. Natl. Acad. Sci. U.S.A. 91 (13): 6098–102. doi:10.1073/pnas.91.13.6098. PMC 44145. PMID 8016121. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=44145.
- ^ Tomasini, Richard; Samir Amina Azizi, Carrier Alice, Isnardon Daniel, Cecchinelli Barbara, Soddu Silvia, Malissen Bernard, Dagorn Jean-Charles, Iovanna Juan L, Dusetti Nelson J (Sep. 2003). "TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity". J. Biol. Chem. 278 (39): 37722–9. doi:10.1074/jbc.M301979200. PMID 12851404.
- ^ Li, L; Liao J, Ruland J, Mak T W, Cohen S N (Feb. 2001). "A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control". Proc. Natl. Acad. Sci. U.S.A. 98 (4): 1619–24. doi:10.1073/pnas.98.4.1619. PMC 29306. PMID 11172000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=29306.
- ^ Lyakhovich, Alex; Shekhar Malathy P V (Apr. 2003). "Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response". Mol. Cell. Biol. 23 (7): 2463–75. doi:10.1128/MCB.23.7.2463-2475.2003. PMC 150718. PMID 12640129. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=150718.
- ^ Li, Muyang; Chen Delin, Shiloh Ariel, Luo Jianyuan, Nikolaev Anatoly Y, Qin Jun, Gu Wei (Apr. 2002). "Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization". Nature 416 (6881): 648–53. doi:10.1038/nature737. PMID 11923872.
- ^ Yang, Qin; Zhang Ran, Wang Xin Wei, Spillare Elisa A, Linke Steven P, Subramanian Deepa, Griffith Jack D, Li Ji Liang, Hickson Ian D, Shen Jiang Cheng, Loeb Lawrence A, Mazur Sharlyn J, Appella Ettore, Brosh Robert M, Karmakar Parimal, Bohr Vilhelm A, Harris Curtis C (Aug. 2002). "The processing of Holliday junctions by BLM and WRN helicases is regulated by p53". J. Biol. Chem. 277 (35): 31980–7. doi:10.1074/jbc.M204111200. PMID 12080066.
- ^ Chang, N S; Pratt N, Heath J, Schultz L, Sleve D, Carey G B, Zevotek N (Feb. 2001). "Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity". J. Biol. Chem. 276 (5): 3361–70. doi:10.1074/jbc.M007140200. PMID 11058590.
- ^ Kelley, K. D.; Miller, K. R.; Todd, A.; Kelley, A. R.; Tuttle, R.; Berberich, S. J. (2010). "YPEL3, a p53-Regulated Gene that Induces Cellular Senescence". Cancer Research 70 (9): 3566–75. doi:10.1158/0008-5472.CAN-09-3219. PMC 2862112. PMID 20388804. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2862112.
- ^ Waterman, M J; Stavridi E S, Waterman J L, Halazonetis T D (Jun. 1998). "ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins". Nat. Genet. 19 (2): 175–8. doi:10.1038/542. PMID 9620776.
- ^ Liu, J; Grogan L, Nau M M, Allegra C J, Chu E, Wright J J (Apr. 2001). "Physical interaction between p53 and primary response gene Egr-1". Int. J. Oncol. 18 (4): 863–70. PMID 11251186.
- ^ Bai, L; Merchant J L (Jul. 2001). "ZBP-89 promotes growth arrest through stabilization of p53". Mol. Cell. Biol. 21 (14): 4670–83. doi:10.1128/MCB.21.14.4670-4683.2001. PMC 87140. PMID 11416144. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=87140.
External links
- a humorous song describing p53 structure and function
- "p53 Knowledgebase". Lane Group at the Institute of Molecular and Cell Biology (IMCB), Singapore. http://p53.bii.a-star.edu.sg/. Retrieved 2008-04-06.
- GeneReviews/NCBI/NIH/UW entry on Li-Fraumeni Syndrome
- TUMOR PROTEIN p53 @ OMIM
- p53 @ The Atlas of Genetics and Cytogenetics in Oncology and Haematology
- TP53 Gene @ GeneCards
- p53 News provided by insciences organisation
- David S. Goodsell (2002-07-01). "p53 Tumor Suppressor". Molecule of the Month. RCSB Protein Data Bank. http://www.pdb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb31_2.html. Retrieved 2008-04-06.
- Thierry Soussi. "p53 Web Site". http://p53.free.fr/. Retrieved 2008-04-06.
- IARC TP53 Somatic Mutations database maintained at IARC, Lyon, by Magali Olivier
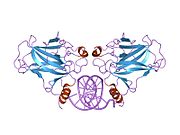
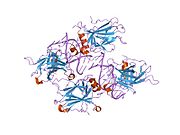
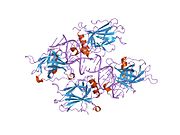
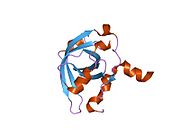
PDB gallery 1a1u: SOLUTION STRUCTURE DETERMINATION OF A P53 MUTANT DIMERIZATION DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE1aie: P53 TETRAMERIZATION DOMAIN CRYSTAL STRUCTURE1c26: CRYSTAL STRUCTURE OF P53 TETRAMERIZATION DOMAIN1gzh: CRYSTAL STRUCTURE OF THE BRCT DOMAINS OF HUMAN 53BP1 BOUND TO THE P53 TUMOR SUPRESSOR1hs5: NMR SOLUTION STRUCTURE OF DESIGNED P53 DIMER1kzy: Crystal Structure of the 53bp1 BRCT Region Complexed to Tumor Suppressor P531olg: HIGH-RESOLUTION SOLUTION STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR1olh: HIGH-RESOLUTION SOLUTION STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR1pes: NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P531pet: NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P531sae: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAC STRUCTURES)1saf: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAD STRUCTURES)1sag:1sah: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAD STRUCTURES)1sai:1saj: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAD STRUCTURES)1sak: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAC STRUCTURES)1sal: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAD STRUCTURES)1tsr: P53 CORE DOMAIN IN COMPLEX WITH DNA1tup: TUMOR SUPPRESSOR P53 COMPLEXED WITH DNA1uol: CRYSTAL STRUCTURE OF THE HUMAN P53 CORE DOMAIN MUTANT M133L/V203A/N239Y/N268D AT 1.9 A RESOLUTION.1ycs: P53-53BP2 COMPLEX2ac0: Structural Basis of DNA Recognition by p53 Tetramers (complex I)2ady: Structural Basis of DNA Recognition by p53 Tetramers (complex IV)2ahi: Structural Basis of DNA Recognition by p53 Tetramers (complex III)2ata: Structural Basis of DNA Recognition by p53 Tetramers (complex II)2b3g: p53N (fragment 33-60) bound to RPA70N2bim: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-N239Y-N268D-R273H2bin: HUMAN P53 CORE DOMAIN MUTANT M133L-H168R-V203A-N239Y-N268D2bio: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-N239Y-R249S-N268D2bip: HUMAN P53 CORE DOMAIN MUTANT M133L-H168R-V203A-N239Y-R249S-N268D2biq: HUMAN P53 CORE DOMAIN MUTANT T123A-M133L-H168R-V203A-N239Y-R249S-N268D2fej: Solution structure of human p53 DNA binding domain.2gs0: NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the activation domain of p532h1l: The Structure of the Oncoprotein SV40 Large T Antigen and p53 Tumor Suppressor Complex2j1w: HUMAN P53 CORE DOMAIN MUTANT M133L-V143A-V203A-N239Y-N268D2j1x: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-Y220C-N239Y-N268D2j1y: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-N239Y-G245S-N268D2j1z: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-N239Y-N268D-F270L2j20: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-N239Y-N268D-R273C2j21: HUMAN P53 CORE DOMAIN MUTANT M133L-V203A-N239Y-N268D-R282W2ocj: Human p53 core domain in the absence of DNA3sak: HIGH RESOLUTION SOLUTION NMR STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF P53 BY MULTI-DIMENSIONAL NMR (SAC STRUCTURES)Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) MiscellaneousNeoplasm: Tumor suppressor genes/proteins and Oncogenes/Proto-oncogenes Ligand Receptor TSP: CDH1TSP: PTCH1TSP: TGF beta receptor 2Intracellular signaling P+Ps ONCO: Beta-catenin · TSP: APCHippo signaling pathwayOther/unknownNucleus TSP: VHL · ONCO: CBL - MDM2Mitochondria Other/ungrouped M: NEO
tsoc, mrkr
tumr, epon, para
drug (L1i/1e/V03)
Cell cycle proteins Cyclin CDK CDK inhibitor P53 p63 p73 family Phases and
checkpointsOther cellular phasesCategories:- Human proteins
- Programmed cell death
- Proteins
- Transcription factors
- Tumor suppressor genes
- Apoptosis
Wikimedia Foundation. 2010.