- Receptor (biochemistry)
-
For other uses, see Receptor (disambiguation).
In biochemistry, a receptor is a molecule found on the surface of a cell, which receives specific chemical signals from neighbouring cells or the wider environment within an organism. These signals tell a cell to do something—for example to divide or die, or to allow certain molecules to enter or exit the cell.
Receptors are protein molecules, embedded in either the plasma membrane (cell surface receptors) or the cytoplasm (nuclear receptors) of a cell, to which one or more specific kinds of signaling molecules may attach. A molecule which binds (attaches) to a receptor is called a ligand, and may be a peptide (short protein) or other small molecule, such as a neurotransmitter, a hormone, a pharmaceutical drug, or a toxin. Each kind of receptor can bind only certain ligand shapes. Each cell typically has many receptors, of many different kinds. Simply put, a receptor functions as a keyhole that opens a biochemical pathway when the proper ligand is inserted.
Ligand binding stabilizes a certain receptor conformation (the three-dimensional shape of the receptor protein, with no change in sequence). This is often associated with gain of or loss of protein activity, ordinarily leading to some sort of cellular response. However, some ligands (e.g. antagonists) merely block receptors without inducing any response. Ligand-induced changes in receptors result in cellular changes which constitute the biological activity of the ligands. Many functions of the human body are regulated by these receptors responding uniquely to specific molecules like this.
Contents
Overview
The shapes and actions of receptors are studied by X-ray crystallography, dual polarisation interferometry, computer modelling, and structure-function studies, which have advanced the understanding of drug action at the binding sites of receptors. Structure activity relationships correlate induced conformational changes with biomolecular activity, and are studied using dynamic techniques such as circular dichroism and dual polarisation interferometry.
Depending on their functions and ligands, several types of receptors may be identified:
- Some receptor proteins are peripheral membrane proteins.
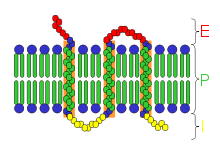
- Many hormone and neurotransmitter receptors are transmembrane proteins: transmembrane receptors are embedded in the phospholipid bilayer of cell membranes, that allow the activation of signal transduction pathways in response to the activation by the binding molecule, or ligand.
- Metabotropic receptors are coupled to G proteins and affect the cell indirectly through enzymes which control ion channels.
- Ionotropic receptors (also known as ligand-gated ion channels) contain a central pore which opens in response to the binding of ligand.
- Another major class of receptors are intracellular proteins such as those for steroid and intracrine peptide hormone receptors. These receptors often can enter the cell nucleus and modulate gene expression in response to the activation by the ligand.
Membrane receptors are isolated from cell membranes by complex extraction procedures using solvents, detergents, and/or affinity purification.
Binding and activation
Ligand binding is an equilibrium process. Ligands bind to receptors and dissociate from them according to the law of mass action.
![\left[\mathrm{Ligand}\right] \cdot \left[\mathrm{Receptor}\right]\;\;\overset{K_d}{\rightleftharpoons}\;\;\left[\text{Ligand-receptor complex}\right]](7/9f79a27e10544f31ae96f42ec21fe0cb.png)
- (the brackets stand for concentrations)
One measure of how well a molecule fits a receptor is the binding affinity, which is inversely related to the dissociation constant Kd. A good fit corresponds with high affinity and low Kd. The final biological response (e.g. second messenger cascade, muscle contraction), is only achieved after a significant number of receptors are activated.
The receptor-ligand affinity is greater than enzyme-substrate affinity.[citation needed] Whilst both interactions are specific and reversible, there is no chemical modification of the ligand as seen with the substrate upon binding to its enzyme.
If the receptor exists in two states (see this picture), then the ligand binding must account for these two receptor states. For a more detailed discussion of two-state binding, which is thought to occur as an activation mechanism in many receptors see this link.
Constitutive activity
A receptor which is capable of producing its biological response in the absence of a bound ligand is said to display "constitutive activity".[1] The constitutive activity of receptors may be blocked by inverse agonist binding. Mutations in receptors that result in increased constitutive activity underlie some inherited diseases, such as precocious puberty (due to mutations in luteinizing hormone receptors) and hyperthyroidism (due to mutations in thyroid-stimulating hormone receptors). For the use of statistical mechanics in a quantitative study of the ligand-receptor binding affinity, see the comprehensive article[2] on the configuration integral.
Theories of drug receptor interaction
Occupation theory
Drug effect is directly proportional to number of receptors occupied. Drug effect ceases as drug-receptor complex dissociate
Ariens & Stephenson theory
introduced Terms of "affinity" & "efficacy" Affinity: ability of the drug to combine with receptor to create drug-receptor complex Efficacy: ability of the drug-receptor complex to initiate a response
Affinity “drug-receptor interaction” is governed by the law of mass action.
In this theory
Agonist: drug with high affinity & high intrinsic activity Partial agonist: drug with high affinity & low intrinsic activity Antagonist: drug with high affinity & low intrinsic activity
Rate theory
The activation of receptors is directly proportional to the total number of encounters of the drug with its receptors per unit time Pharmacological activity is directly proportional to the rate of dissociation & association not number of receptors occupied
Agonist:drug with fast association & fast dissociation Partial agonist:drug with intermediate association & intermediate dissociation Antagonist:drug with fast association & slow dissociation
Induced fit theory
As the drug approaches the receptor the receptor alters the conformation of its binding site to produce drug—receptor complex
Agonists versus antagonists
Not every ligand that binds to a receptor also activates the receptor. The following classes of ligands exist:
- (Full) agonists are able to activate the receptor and result in a maximal biological response. Most natural ligands are full agonists.
- Partial agonists do not activate receptors thoroughly, causing responses which are partial compared to those of full agonists.
- Antagonists bind to receptors but do not activate them. This results in receptor blockage, inhibiting the binding of other agonists.
- Inverse agonists reduce the activity of receptors by inhibiting their constitutive activity.
Peripheral membrane protein receptors
These receptors are relatively rare compared to the much more common types of receptors that cross the cell membrane. An example of a receptor that is a peripheral membrane protein is the elastin receptor.
Transmembrane receptors
Metabotropic receptors
G protein-coupled receptors
These receptors are also known as seven transmembrane receptors or 7TM receptors, because they pass through the membrane seven times.
- Muscarinic acetylcholine receptor (Acetylcholine and Muscarine)
- Adenosine receptors (Adenosine)
- Adrenoceptors (also known as Adrenergic receptors, for adrenaline, and other structurally related hormones and drugs)
- GABA receptors, Type-B (γ-Aminobutyric acid or GABA)
- Angiotensin receptors (Angiotensin)
- Cannabinoid receptors (Cannabinoids)
- Cholecystokinin receptors (Cholecystokinin)
- Dopamine receptors (Dopamine)
- Glucagon receptors (Glucagon)
- Melatonin receptors (Melatonin)
- Metabotropic glutamate receptors (Glutamate)
- Histamine receptors (Histamine)
- Olfactory receptors (for the sense of smell)
- Opioid receptors (Opioids)
- Protease-activated receptors
- Rhodopsin (a photoreceptor protein)
- Secretin receptors (Secretin)
- Serotonin receptors, except Type-3 (Serotonin, also known as 5-Hydroxytryptamine or 5-HT)
- Somatostatin receptors (Somatostatin)
- Trace-amine associated receptors (Trace amines)
- Calcium-sensing receptor (Calcium)
- Chemokine receptors (Chemokines)
- many more ...
Receptor tyrosine kinases
These receptors detect ligands and propagate signals via the tyrosine kinase of their intracellular domains. This family of receptors includes;
- Erythropoietin receptor (Erythropoietin)
- Insulin receptor (Insulin)
- Eph receptors
- Insulin-like growth factor 1 receptor
- various other growth factor and cytokine receptors
- ....
Guanylyl cyclase receptors
- GC-A & GC-B: receptors for Atrial-natriuretic peptide (ANP) and other natriuretic peptides
- GC-C: Guanylin receptor
Ionotropic receptors
Main article: Ionotropic receptorIonotropic receptors are heteromeric or homomeric oligomers.[3] They are receptors that respond to extracellular ligands and receptors that respond to intracellular ligands.
Extracellular ligands
Receptor Ligand Ion current Nicotinic acetylcholine receptor Acetylcholine, Nicotine Na+, K+, Ca2+ [3] Glycine receptor (GlyR) Glycine, Strychnine Cl− > HCO−3 [3] GABA receptors: GABA-A, GABA-C GABA Cl− > HCO−3 [3] Glutamate receptors: NMDA receptor, AMPA receptor, and Kainate receptor Glutamate Na+, K+, Ca2+ [3] 5-HT3 receptor Serotonin Na+, K+ [3] P2X receptors ATP Ca2+, Na+, Mg2+ [3] Intracellular ligands
Receptor Ligand Ion current cyclic nucleotide-gated ion channels cGMP (vision), cAMP and cGTP (olfaction) Na+, K+ [3] IP3 receptor IP3 Ca2+ [3] Intracellular ATP receptors ATP (closes channel)[3] K+ [3] Ryanodine receptor Ca2+ Ca2+ [3] The entire repertoire of human plasma membrane receptors is listed at the Human Plasma Membrane Receptome (http://www.receptome.org).
Intracellular receptors
Transcription factors
Various
- Ionotropic receptors (IP3 receptor above)
- sigma1 (neurosteroids)
- G protein-coupled receptors [4]
Role in genetic disorders
Many genetic disorders involve hereditary defects in receptor genes. Often, it is hard to determine whether the receptor is nonfunctional or the hormone is produced at decreased level; this gives rise to the "pseudo-hypo-" group of endocrine disorders, where there appears to be a decreased hormonal level while in fact it is the receptor that is not responding sufficiently to the hormone.
Receptor regulation
Cells can increase (upregulate) or decrease (downregulate) the number of receptors to a given hormone or neurotransmitter to alter its sensitivity to this molecule. This is a locally acting feedback mechanism.
- Receptor desensitization[5][broken citation]
- Uncoupling of receptor effector molecules.
- Receptor sequestration (internalization).[6]
In immune system
The main receptors in the immune system are pattern recognition receptors (PRRs), toll-like receptors (TLRs), killer activated and killer inhibitor receptors (KARs and KIRs), complement receptors, Fc receptors, B cell receptors and T cell receptors.[7]
Transmembrane receptors: Immunoglobulin superfamily immune receptors Antibody receptor:
Fc receptorSecretoryAntigen receptor Antigen receptorAccessory moleculesT cellsAntigen receptorAccessory moleculesCytokine receptor see cytokine receptorsKiller-cell IG-like receptors Leukocyte IG-like receptors B trdu: iter (nrpl/grfl/cytl/horl), csrc (lgic, enzr, gprc, igsr, intg, nrpr/grfr/cytr), itra (adap, gbpr, mapk), calc, lipd; path (hedp, wntp, tgfp+mapp, notp, jakp, fsap, hipp, tlrp)See also
- Ki Database
- Ion channel linked receptors
- Neuropsychopharmacology
- Schild regression for ligand receptor inhibition
- Signal transduction
- Stem cell marker
References
- ^ Milligan G (December 2003). "Constitutive activity and inverse agonists of G protein-coupled receptors: a current perspective". Mol. Pharmacol. 64 (6): 1271–6. doi:10.1124/mol.64.6.1271. PMID 14645655.
- ^ Vu-Quoc, L., Configuration integral (statistical mechanics), 2008.
- ^ a b c d e f g h i j k l Medical Physiology, Boron & Boulpaep, ISBN 1-4160-2328-3, Elsevier Saunders 2005. Updated edition. Page 90.
- ^ Gobeil F, et al. (2006) G-protein-coupled receptors signalling at the cell nucleus: an emerging paradigm. Can J Physiol Pharmacol. 2006 Mar–Apr;84(3–4):287–97. PMID 16902576
- ^ Ligand-bound desensitation Vol. 135. No. 5 2130–2136
- ^ G. Boulay, L. Chrbtien, D.E. Richard, AND G. Guillemettes. (1994) Short-Term Desensitization of the Angiotensin II Receptor of Bovine Adrenal Glomerulosa Cells Corresponds to a Shift from a High to a Low Affinity State. Endocrinology Vol. 135. No. 5 2130–2136
- ^ Lippincott's Illustrated Reviews: Immunology. Paperback: 384 pages. Publisher: Lippincott Williams & Wilkins; (July 1, 2007). Language: English. ISBN 0-7817-9543-5. ISBN 978-0-7817-9543-2. Page 20
External links
- IUPHAR GPCR Database and Ion Channels Compendium
- Human plasma membrane receptome
- MeSH Cell+surface+receptors
Signaling pathways Agents ReceptorsIntracellular signaling P+PsSignal transducing adaptor protein: Scaffold protein
2nd messenger: cAMP-dependent pathway · Ca2+ signaling · Lipid signaling · IP3/DAG pathwayBy location Other concepts Membrane proteins, receptors: cell surface receptors G protein-coupled receptor Class AClass BClass CClass DPheromone receptorClass EcAMP receptorClass FLigand-gated ion channel Enzyme-linked receptor Other/ungrouped Asialoglycoprotein receptor · Tumor necrosis factor receptor · Immunoglobulin superfamily · N-Acetylglucosamine receptor · Neuropilins · Transferrin receptor · EDAR · Lipoprotein receptor-related proteinTranscription factors and intracellular receptors (1) Basic domains Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains ARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.3) p53(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.6) MiscellaneousCategories:
Wikimedia Foundation. 2010.