- Liver X receptor alpha
-
Liver X receptor alpha (LXR-alpha) is a nuclear receptor protein that in humans is encoded by the NR1H3 gene (nuclear receptor subfamily 1, group H, member 3).[1][2]
Contents
Expression
miRNA hsa-miR-613 autoregulates the human LXRα gene by targeting the endogenous LXRα through its specific miRNA response element (613MRE) within the LXRα 3′-untranslated region. LXRα autoregulates its own suppression via induction of SREBP1c which upregulates miRNA hsa-miR-613.[3]
Function
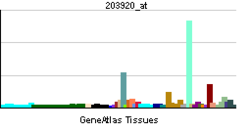
The liver X receptors, LXRα (this protein) and LXRβ, form a subfamily of the nuclear receptor superfamily and are key regulators of macrophage function, controlling transcriptional programs involved in lipid homeostasis and inflammation. The inducible LXRα is highly expressed in liver, adrenal gland, intestine, adipose tissue, macrophages, lung, and kidney, whereas LXRβ is ubiquitously expressed. Ligand-activated LXRs form obligate heterodimers with retinoid X receptors (RXRs) and regulate expression of target genes containing LXR response elements.[4][5]
Interactions
Liver X receptor alpha has been shown to interact with EDF1[6] and Small heterodimer partner.[7]
References
- ^ Miyata KS, McCaw SE, Patel HV, Rachubinski RA, Capone JP (1996). "The orphan nuclear hormone receptor LXR alpha interacts with the peroxisome proliferator-activated receptor and inhibits peroxisome proliferator signaling". J. Biol. Chem. 271 (16): 9189–92. doi:10.1074/jbc.271.16.9189. PMID 8621574.
- ^ Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ (1995). "LXR, a nuclear receptor that defines a distinct retinoid response pathway". Genes Dev. 9 (9): 1033–45. doi:10.1101/gad.9.9.1033. PMID 7744246.
- ^ http://mend.endojournals.org/content/25/4/584.abstract
- ^ Korf H, Vander Beken S, Romano M, Steffensen KR, Stijlemans B, Gustafsson JA, Grooten J, Huygen K (June 2009). "Liver X receptors contribute to the protective immune response against Mycobacterium tuberculosis in mice". J. Clin. Invest. 119 (6): 1626–37. doi:10.1172/JCI35288. PMC 2689129. PMID 19436111. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2689129.
- ^ "Entrez Gene: nuclear receptor subfamily 1". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=10062.
- ^ Brendel C, Gelman L, Auwerx J (June 2002). "Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism". Mol. Endocrinol. 16 (6): 1367–77. doi:10.1210/me.16.6.1367. PMID 12040021.
- ^ Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J (September 2002). "The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity". Mol. Endocrinol. 16 (9): 2065–76. doi:10.1210/me.2001-0194. PMID 12198243.
Further reading
- Lehmann JM, Kliewer SA, Moore LB, et al. (1997). "Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway.". J. Biol. Chem. 272 (6): 3137–40. doi:10.1074/jbc.272.6.3137. PMID 9013544.
- Auboeuf D, Rieusset J, Fajas L, et al. (1997). "Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-alpha in humans: no alteration in adipose tissue of obese and NIDDM patients.". Diabetes 46 (8): 1319–27. doi:10.2337/diabetes.46.8.1319. PMID 9231657.
- Peet DJ, Turley SD, Ma W, et al. (1998). "Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha.". Cell 93 (5): 693–704. doi:10.1016/S0092-8674(00)81432-4. PMID 9630215.
- Miyata KS, McCaw SE, Meertens LM, et al. (1999). "Receptor-interacting protein 140 interacts with and inhibits transactivation by, peroxisome proliferator-activated receptor alpha and liver-X-receptor alpha.". Mol. Cell. Endocrinol. 146 (1-2): 69–76. doi:10.1016/S0303-7207(98)00196-8. PMID 10022764.
- Venkateswaran A, Laffitte BA, Joseph SB, et al. (2000). "Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha.". Proc. Natl. Acad. Sci. U.S.A. 97 (22): 12097–102. doi:10.1073/pnas.200367697. PMC 17300. PMID 11035776. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=17300.
- Chinetti G, Lestavel S, Bocher V, et al. (2001). "PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway.". Nat. Med. 7 (1): 53–8. doi:10.1038/83348. PMID 11135616.
- Laffitte BA, Repa JJ, Joseph SB, et al. (2001). "LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes.". Proc. Natl. Acad. Sci. U.S.A. 98 (2): 507–12. doi:10.1073/pnas.021488798. PMC 14617. PMID 11149950. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=14617.
- Li Y, Bolten C, Bhat BG, et al. (2002). "Induction of human liver X receptor alpha gene expression via an autoregulatory loop mechanism.". Mol. Endocrinol. 16 (3): 506–14. doi:10.1210/me.16.3.506. PMID 11875109.
- Mak PA, Laffitte BA, Desrumaux C, et al. (2002). "Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta.". J. Biol. Chem. 277 (35): 31900–8. doi:10.1074/jbc.M202993200. PMID 12032151.
- Brendel C, Gelman L, Auwerx J (2003). "Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism.". Mol. Endocrinol. 16 (6): 1367–77. doi:10.1210/me.16.6.1367. PMID 12040021.
- Steffensen KR, Schuster GU, Parini P, et al. (2002). "Different regulation of the LXRalpha promoter activity by isoforms of CCAAT/enhancer-binding proteins.". Biochem. Biophys. Res. Commun. 293 (5): 1333–40. doi:10.1016/S0006-291X(02)00390-X. PMID 12054659.
- Gbaguidi GF, Agellon LB (2002). "The atypical interaction of peroxisome proliferator-activated receptor alpha with liver X receptor alpha antagonizes the stimulatory effect of their respective ligands on the murine cholesterol 7alpha-hydroxylase gene promoter.". Biochim. Biophys. Acta 1583 (2): 229–36. PMID 12117567.
- Pawar A, Xu J, Jerks E, et al. (2002). "Fatty acid regulation of liver X receptors (LXR) and peroxisome proliferator-activated receptor alpha (PPARalpha ) in HEK293 cells.". J. Biol. Chem. 277 (42): 39243–50. doi:10.1074/jbc.M206170200. PMID 12161442.
- Brendel C, Schoonjans K, Botrugno OA, et al. (2003). "The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity.". Mol. Endocrinol. 16 (9): 2065–76. doi:10.1210/me.2001-0194. PMID 12198243.
- Oberkofler H, Schraml E, Krempler F, Patsch W (2003). "Potentiation of liver X receptor transcriptional activity by peroxisome-proliferator-activated receptor gamma co-activator 1 alpha.". Biochem. J. 371 (Pt 1): 89–96. doi:10.1042/BJ20021665. PMC 1223253. PMID 12470296. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1223253.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=139241.
- Wang M, Thomas J, Burris TP, et al. (2004). "Molecular determinants of LXRalpha agonism.". J. Mol. Graph. Model. 22 (2): 173–81. doi:10.1016/S1093-3263(03)00159-1. PMID 12932788.
- Fukuchi J, Song C, Ko AL, Liao S (2004). "Transcriptional regulation of farnesyl pyrophosphate synthase by liver X receptors.". Steroids 68 (7-8): 685–91. doi:10.1016/S0039-128X(03)00100-4. PMID 12957674.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
PDB gallery Transcription factors and intracellular receptors (1) Basic domains Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains ARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.3) p53(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.6) MiscellaneousCategories:- Human proteins
- Chromosome 11 gene stubs
- Intracellular receptors
- Transcription factors
Wikimedia Foundation. 2010.