- DIDO1
-
Death inducer-obliterator 1 
PDB rendering based on 1wem.Available structures PDB 1wem Identifiers Symbols DIDO1; BYE1; C20orf158; DATF1; DIDO2; DIDO3; DIO-1; DIO1; DKFZp434P1115; FLJ11265; KIAA0333; MGC16140; dJ885L7.8 External IDs OMIM: 604140 MGI: 1344352 HomoloGene: 34139 GeneCards: DIDO1 Gene Gene Ontology Molecular function • zinc ion binding
• metal ion bindingCellular component • nucleus
• cytoplasmBiological process • transcription, DNA-dependent
• apoptosisSources: Amigo / QuickGO RNA expression pattern 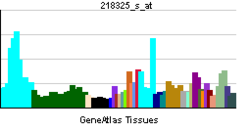
More reference expression data Orthologs Species Human Mouse Entrez 11083 23856 Ensembl ENSG00000101191 ENSMUSG00000038914 UniProt Q9BTC0 Q05C59 RefSeq (mRNA) NM_001193369.1 NM_011805 RefSeq (protein) NP_001180298.1 NP_035935 Location (UCSC) Chr 20:
61.51 – 61.57 MbChr 2:
180.39 – 180.44 MbPubMed search [1] [2] Death-inducer obliterator 1 is a protein that in humans is encoded by the DIDO1 gene.[1][2]
Contents
Function
Apoptosis, a major form of cell death, is an efficient mechanism for eliminating unwanted cells and is of central importance for development and homeostasis in metazoan animals. In mice, the death inducer-obliterator-1 gene is upregulated by apoptotic signals and encodes a cytoplasmic protein that translocates to the nucleus upon apoptotic signal activation. When overexpressed, the mouse protein induced apoptosis in cell lines growing in vitro. This gene is similar to the mouse gene and therefore is thought to be involved in apoptosis. Alternatively spliced transcripts have been found for this gene, encoding multiple isoforms.[2]
References
- ^ Garcia-Domingo D, Leonardo E, Grandien A, Martinez P, Albar JP, Izpisua-Belmonte JC, Martinez-A C (Aug 1999). "DIO-1 is a gene involved in onset of apoptosis in vitro, whose misexpression disrupts limb development". Proc Natl Acad Sci U S A 96 (14): 7992–7. doi:10.1073/pnas.96.14.7992. PMC 22175. PMID 10393935. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=22175.
- ^ a b "Entrez Gene: DIDO1 death inducer-obliterator 1". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=11083.
Further reading
- Bonaldo MF, Lennon G, Soares MB (1997). "Normalization and subtraction: two approaches to facilitate gene discovery.". Genome Res. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Nagase T, Ishikawa K, Nakajima D, et al. (1997). "Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro.". DNA Res. 4 (2): 141–50. doi:10.1093/dnares/4.2.141. PMID 9205841.
- Deloukas P, Matthews LH, Ashurst J, et al. (2002). "The DNA sequence and comparative analysis of human chromosome 20.". Nature 414 (6866): 865–71. doi:10.1038/414865a. PMID 11780052.
- Gomes I, Sharma TT, Edassery S, et al. (2002). "Novel transcription factors in human CD34 antigen-positive hematopoietic cells.". Blood 100 (1): 107–19. doi:10.1182/blood.V100.1.107. PMID 12070015.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=139241.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs.". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Brandenberger R, Wei H, Zhang S, et al. (2005). "Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation.". Nat. Biotechnol. 22 (6): 707–16. doi:10.1038/nbt971. PMID 15146197.
- Fu GK, Wang JT, Yang J, et al. (2005). "Circular rapid amplification of cDNA ends for high-throughput extension cloning of partial genes.". Genomics 84 (1): 205–10. doi:10.1016/j.ygeno.2004.01.011. PMID 15203218.
- Beausoleil SA, Jedrychowski M, Schwartz D, et al. (2004). "Large-scale characterization of HeLa cell nuclear phosphoproteins.". Proc. Natl. Acad. Sci. U.S.A. 101 (33): 12130–5. doi:10.1073/pnas.0404720101. PMC 514446. PMID 15302935. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=514446.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC).". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=528928.
- Fütterer A, Campanero MR, Leonardo E, et al. (2005). "Dido gene expression alterations are implicated in the induction of hematological myeloid neoplasms.". J. Clin. Invest. 115 (9): 2351–62. doi:10.1172/JCI24177. PMC 1190370. PMID 16127461. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1190370.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network.". Nature 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Beausoleil SA, Villén J, Gerber SA, et al. (2006). "A probability-based approach for high-throughput protein phosphorylation analysis and site localization.". Nat. Biotechnol. 24 (10): 1285–92. doi:10.1038/nbt1240. PMID 16964243.
- Olsen JV, Blagoev B, Gnad F, et al. (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks.". Cell 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.
- Trachana V, van Wely KH, Guerrero AA, et al. (2007). "Dido disruption leads to centrosome amplification and mitotic checkpoint defects compromising chromosome stability.". Proc. Natl. Acad. Sci. U.S.A. 104 (8): 2691–6. doi:10.1073/pnas.0611132104. PMC 1815243. PMID 17299043. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1815243.
External links
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
PDB gallery Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) Miscellaneoussee also transcription factor/coregulator deficiencies
B bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°)Categories:- Human proteins
- Chromosome 20 gene stubs
- Transcription factors
Wikimedia Foundation. 2010.