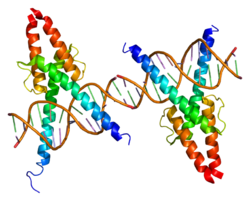
- MyoD
-
MyoD is a protein with a key role in regulating muscle differentiation. MyoD belongs to a family of proteins known as myogenic regulatory factors (MRFs).[1] These bHLH (basic helix loop helix) transcription factors act sequentially in myogenic differentiation. MRF family members include MyoD, Myf5, myogenin, and MRF4 (Myf6).
MyoD is one of the earliest markers of myogenic commitment. MyoD is expressed in activated satellite cells, but not in quiescent satellite cells. Although MyoD marks myoblast commitment, muscle development is not dramatically ablated in mouse mutants lacking the MyoD gene. This is likely to be due to functional redundancy from Myf5.
Contents
Functions of MyoD
The function of MyoD in development is to commit mesoderm cells to a skeletal lineage, and then to regulate that process. MyoD may also play a role in regulating muscle repair. MyoD mRNA levels are also reported to be elevated in aging skeletal muscle.
One of the main actions of MyoD is to remove cells from the cell cycle (halt proliferation) by enhancing the transcription of p21. MyoD is inhibited by cyclin dependent kinases (CDKs). CDKs are in turn inhibited by p21. Thus MyoD enhances its own activity in the cell.
Interactions
MyoD has been shown to interact with HDAC1,[2][3] ID1,[4][5][6][7][8][9] ID2,[5] Retinoblastoma protein,[3][10] STAT3,[11] CSRP3,[12] Cyclin-dependent kinase 4,[13][14] CREB-binding protein,[15][16] TCF3,[5][17] MOS,[18] EP300,[16][19] C-jun,[20] Cyclin-dependent kinase inhibitor 1C,[21] Retinoid X receptor alpha[22] and MDFI.[23]
Coactivators of MyoD
IFRD1 is a positive cofactor of MyoD, as it cooperates with MyoD at inducing the transcriptional activity of MEF2C (by displacing HDAC4 from MEF2C); moreover IFRD1 also represses the transcriptional activity of NF-kB, which is known to inhibit MyoD mRNA accumulation.[24][25]
References
- ^ "Entrez Gene: MYOD1 myogenic differentiation 1". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4654.
- ^ Puri, P L; Iezzi S, Stiegler P, Chen T T, Schiltz R L, Muscat G E, Giordano A, Kedes L, Wang J Y, Sartorelli V (Oct. 2001). "Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis". Mol. Cell (United States) 8 (4): 885–97. doi:10.1016/S1097-2765(01)00373-2. ISSN 1097-2765. PMID 11684023.
- ^ a b Mal, A; Sturniolo M, Schiltz R L, Ghosh M K, Harter M L (Apr. 2001). "A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program". EMBO J. (England) 20 (7): 1739–53. doi:10.1093/emboj/20.7.1739. ISSN 0261-4189. PMC 145490. PMID 11285237. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=145490.
- ^ Garkavtsev, Igor; Kozin Sergey V, Chernova Olga, Xu Lei, Winkler Frank, Brown Edward, Barnett Gene H, Jain Rakesh K (Mar. 2004). "The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis". Nature (England) 428 (6980): 328–32. doi:10.1038/nature02329. PMID 15029197.
- ^ a b c Langlands, K; Yin X, Anand G, Prochownik E V (Aug. 1997). "Differential interactions of Id proteins with basic-helix-loop-helix transcription factors". J. Biol. Chem. (UNITED STATES) 272 (32): 19785–93. doi:10.1074/jbc.272.32.19785. ISSN 0021-9258. PMID 9242638.
- ^ Finkel, T; Duc J, Fearon E R, Dang C V, Tomaselli G F (Jan. 1993). "Detection and modulation in vivo of helix-loop-helix protein-protein interactions". J. Biol. Chem. (UNITED STATES) 268 (1): 5–8. ISSN 0021-9258. PMID 8380166.
- ^ Gupta, K; Anand G, Yin X, Grove L, Prochownik E V (Mar. 1998). "Mmip1: a novel leucine zipper protein that reverses the suppressive effects of Mad family members on c-myc". Oncogene (ENGLAND) 16 (9): 1149–59. doi:10.1038/sj.onc.1201634. ISSN 0950-9232. PMID 9528857.
- ^ McLoughlin, Patricia; Ehler Elisabeth, Carlile Graeme, Licht Jonathan D, Schäfer Beat W (Oct. 2002). "The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein". J. Biol. Chem. (United States) 277 (40): 37045–53. doi:10.1074/jbc.M203336200. ISSN 0021-9258. PMID 12145280.
- ^ Ling, Ming-Tat; Chiu Yung-Tuen, Lee Terence Kin Wah, Leung Steve Chin Lung, Fung Maggie Ka Lai, Wang Xianghong, Wong Kwong Fai, Wong Yong-Chuan (Sep. 2008). "Id-1 induces proteasome-dependent degradation of the HBX protein". J. Mol. Biol. (England) 382 (1): 34–43. doi:10.1016/j.jmb.2007.06.020. PMID 18674781.
- ^ Gu, W; Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B (Feb. 1993). "Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation". Cell (UNITED STATES) 72 (3): 309–24. doi:10.1016/0092-8674(93)90110-C. ISSN 0092-8674. PMID 8381715.
- ^ Kataoka, Yoshihisa; Matsumura Itaru, Ezoe Sachiko, Nakata Soichi, Takigawa Eri, Sato Yusuke, Kawasaki Akira, Yokota Takashi, Nakajima Koichi, Felsani Armando, Kanakura Yuzuru (Nov. 2003). "Reciprocal inhibition between MyoD and STAT3 in the regulation of growth and differentiation of myoblasts". J. Biol. Chem. (United States) 278 (45): 44178–87. doi:10.1074/jbc.M304884200. ISSN 0021-9258. PMID 12947115.
- ^ Kong, Y; Flick M J, Kudla A J, Konieczny S F (Aug. 1997). "Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD". Mol. Cell. Biol. (UNITED STATES) 17 (8): 4750–60. ISSN 0270-7306. PMC 232327. PMID 9234731. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=232327.
- ^ Zhang, J M; Zhao X, Wei Q, Paterson B M (Dec. 1999). "Direct inhibition of G(1) cdk kinase activity by MyoD promotes myoblast cell cycle withdrawal and terminal differentiation". EMBO J. (ENGLAND) 18 (24): 6983–93. doi:10.1093/emboj/18.24.6983. ISSN 0261-4189. PMC 1171761. PMID 10601020. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1171761.
- ^ Zhang, J M; Wei Q, Zhao X, Paterson B M (Feb. 1999). "Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4". EMBO J. (ENGLAND) 18 (4): 926–33. doi:10.1093/emboj/18.4.926. ISSN 0261-4189. PMC 1171185. PMID 10022835. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1171185.
- ^ Polesskaya, A; Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A (Aug. 2001). "Interaction between acetylated MyoD and the bromodomain of CBP and/or p300". Mol. Cell. Biol. (United States) 21 (16): 5312–20. doi:10.1128/MCB.21.16.5312-5320.2001. ISSN 0270-7306. PMC 87255. PMID 11463815. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=87255.
- ^ a b Sartorelli, V; Huang J, Hamamori Y, Kedes L (Feb. 1997). "Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C". Mol. Cell. Biol. (UNITED STATES) 17 (2): 1010–26. ISSN 0270-7306. PMC 231826. PMID 9001254. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=231826.
- ^ Maleki, S J; Royer C A, Hurlburt B K (Jun. 1997). "MyoD-E12 heterodimers and MyoD-MyoD homodimers are equally stable". Biochemistry (UNITED STATES) 36 (22): 6762–7. doi:10.1021/bi970262m. ISSN 0006-2960. PMID 9184158.
- ^ Lenormand, J L; Benayoun B, Guillier M, Vandromme M, Leibovitch M P, Leibovitch S A (Feb. 1997). "Mos activates myogenic differentiation by promoting heterodimerization of MyoD and E12 proteins". Mol. Cell. Biol. (UNITED STATES) 17 (2): 584–93. ISSN 0270-7306. PMC 231783. PMID 9001211. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=231783.
- ^ Lau, P; Bailey P, Dowhan D H, Muscat G E (Jan. 1999). "Exogenous expression of a dominant negative RORalpha1 vector in muscle cells impairs differentiation: RORalpha1 directly interacts with p300 and myoD". Nucleic Acids Res. (ENGLAND) 27 (2): 411–20. doi:10.1093/nar/27.2.411. ISSN 0305-1048. PMC 148194. PMID 9862959. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=148194.
- ^ Bengal, E; Ransone L, Scharfmann R, Dwarki V J, Tapscott S J, Weintraub H, Verma I M (Feb. 1992). "Functional antagonism between c-Jun and MyoD proteins: a direct physical association". Cell (UNITED STATES) 68 (3): 507–19. doi:10.1016/0092-8674(92)90187-H. ISSN 0092-8674. PMID 1310896.
- ^ Reynaud, E G; Leibovitch M P, Tintignac L A, Pelpel K, Guillier M, Leibovitch S A (Jun. 2000). "Stabilization of MyoD by direct binding to p57(Kip2)". J. Biol. Chem. (UNITED STATES) 275 (25): 18767–76. doi:10.1074/jbc.M907412199. ISSN 0021-9258. PMID 10764802.
- ^ Froeschlé, A; Alric S, Kitzmann M, Carnac G, Auradé F, Rochette-Egly C, Bonnieu A (Jul. 1998). "Retinoic acid receptors and muscle b-HLH proteins: partners in retinoid-induced myogenesis". Oncogene (ENGLAND) 16 (26): 3369–78. doi:10.1038/sj.onc.1201894. ISSN 0950-9232. PMID 9692544.
- ^ Chen, C M; Kraut N, Groudine M, Weintraub H (Sep. 1996). "I-mf, a novel myogenic repressor, interacts with members of the MyoD family". Cell (UNITED STATES) 86 (5): 731–41. doi:10.1016/S0092-8674(00)80148-8. ISSN 0092-8674. PMID 8797820.
- ^ Micheli L, Leonardi L, Conti F, Buanne P, Canu N, Caruso M, Tirone F (March 2005). "PC4 coactivates MyoD by relieving the histone deacetylase 4-mediated inhibition of myocyte enhancer factor 2C". Mol. Cell. Biol. 25 (6): 2242–59. doi:10.1128/MCB.25.6.2242-2259.2005. PMC 1061592. PMID 15743821. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1061592.
- ^ Micheli L, Leonardi L, Conti F, Maresca G, Colazingari S, Mattei E, Lira SA, Farioli-Vecchioli S, Caruso M, Tirone F (February 2011). "PC4/Tis7/IFRD1 stimulates skeletal muscle regeneration and is involved in myoblast differentiation as a regulator of MyoD and NF-kappaB". J. Biol. Chem. 286 (7): 5691–707. doi:10.1074/jbc.M110.162842. PMC 3037682. PMID 21127072. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3037682.
Further reading
- Weintraub H, Davis R, Tapscott S et al. (1991). "The myoD gene family: nodal point during specification of the muscle cell lineage". Science 251 (4995): 761–6. doi:10.1126/science.1846704. PMID 1846704.
- Tapscott SJ, Weintraub H (1991). "MyoD and the regulation of myogenesis by helix-loop-helix proteins". J. Clin. Invest. 87 (4): 1133–8. doi:10.1172/JCI115109. PMC 295115. PMID 1849142. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=295115.
- Olson EN (1991). "MyoD family: a paradigm for development?". Genes Dev. 4 (9): 1454–61. doi:10.1101/gad.4.9.1454. PMID 2253873.
- Goldman PS, Tran VK, Goodman RH (1997). "The multifunctional role of the co-activator CBP in transcriptional regulation". Recent Prog. Horm. Res. 52: 103–19; discussion 119–20. PMID 9238849.
- Puri PL, Sartorelli V (2000). "Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications". J. Cell. Physiol. 185 (2): 155–73. doi:10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. PMID 11025438.
- McKinsey TA, Zhang CL, Olson EN (2001). "Control of muscle development by dueling HATs and HDACs". Curr. Opin. Genet. Dev. 11 (5): 497–504. doi:10.1016/S0959-437X(00)00224-0. PMID 11532390.
- Berkes CA, Tapscott SJ (2006). "MyoD and the transcriptional control of myogenesis". Semin. Cell Dev. Biol. 16 (4–5): 585–95. doi:10.1016/j.semcdb.2005.07.006. PMID 16099183.
- Bengal E, Ransone L, Scharfmann R et al. (1992). "Functional antagonism between c-Jun and MyoD proteins: a direct physical association". Cell 68 (3): 507–19. doi:10.1016/0092-8674(92)90187-H. PMID 1310896.
- Walsh K, Gualberto A (1992). "MyoD binds to the guanine tetrad nucleic acid structure". J. Biol. Chem. 267 (19): 13714–8. PMID 1320026.
- Li L, Zhou J, James G et al. (1993). "FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains". Cell 71 (7): 1181–94. doi:10.1016/S0092-8674(05)80066-2. PMID 1335366.
- Shaknovich R, Shue G, Kohtz DS (1992). "Conformational activation of a basic helix-loop-helix protein (MyoD1) by the C-terminal region of murine HSP90 (HSP84)". Mol. Cell. Biol. 12 (11): 5059–68. PMC 360439. PMID 1406681. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=360439.
- Lassar AB, Davis RL, Wright WE et al. (1991). "Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo". Cell 66 (2): 305–15. doi:10.1016/0092-8674(91)90620-E. PMID 1649701.
- Pearson-White SH (1991). "Human MyoD: cDNA and deduced amino acid sequence". Nucleic Acids Res. 19 (5): 1148–1148. doi:10.1093/nar/19.5.1148. PMC 333794. PMID 1850513. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=333794.
- Gessler M, Hameister H, Henry I et al. (1991). "The human MyoD1 (MYF3) gene maps on the short arm of chromosome 11 but is not associated with the WAGR locus or the region for the Beckwith-Wiedemann syndrome". Hum. Genet. 86 (2): 135–8. PMID 2176177.
- Scrable HJ, Johnson DK, Rinchik EM, Cavenee WK (1990). "Rhabdomyosarcoma-associated locus and MYOD1 are syntenic but separate loci on the short arm of human chromosome 11". Proc. Natl. Acad. Sci. U.S.A. 87 (6): 2182–6. doi:10.1073/pnas.87.6.2182. PMC 53650. PMID 2315312. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=53650.
- Lassar AB, Buskin JN, Lockshon D et al. (1989). "MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer". Cell 58 (5): 823–31. doi:10.1016/0092-8674(89)90935-5. PMID 2550138.
- Braun T, Bober E, Buschhausen-Denker G et al. (1990). "Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products". EMBO J. 8 (12): 3617–25. PMC 402043. PMID 2583111. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=402043.
PDB gallery External links
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) MiscellaneousCategories:- Human proteins
- Genes
- Transcription factors
Wikimedia Foundation. 2010.