- Nuclear receptor
-
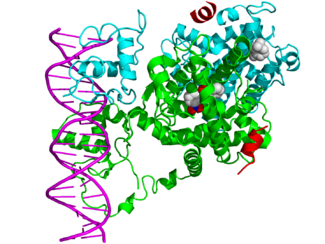 Crystallographic structure of a heterodimer of the nuclear receptors PPAR-γ (green) and RXR-α (cyan) bound to double stranded DNA (magenta) and two molecules of the NCOA2 coactivator (red). The PPAR-γ antagonist GW9662 and RXR-α agonist retinoic acid are depicted as space-filling models (carbon = white, oxygen = red, nitrogen = blue, chlorine = green).[1]
Crystallographic structure of a heterodimer of the nuclear receptors PPAR-γ (green) and RXR-α (cyan) bound to double stranded DNA (magenta) and two molecules of the NCOA2 coactivator (red). The PPAR-γ antagonist GW9662 and RXR-α agonist retinoic acid are depicted as space-filling models (carbon = white, oxygen = red, nitrogen = blue, chlorine = green).[1]
In the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules. In response, these receptors work with other proteins to regulate the expression of specific genes, thereby controlling the development, homeostasis, and metabolism of the organism.
Nuclear receptors have the ability to directly bind to DNA and regulate the expression of adjacent genes, hence these receptors are classified as transcription factors.[2][3] The regulation of gene expression by nuclear receptors generally only happens when a ligand — a molecule that affects the receptor's behavior — is present. More specifically, ligand binding to a nuclear receptor results in a conformational change in the receptor, which, in turn, activates the receptor, resulting in up-regulation or down-regulation of gene expression.
A unique property of nuclear receptors that differentiates them from other classes of receptors is their ability to directly interact with and control the expression of genomic DNA. As a consequence, nuclear receptors play key roles in both embryonic development and adult homeostasis. As discussed below, nuclear receptors may be classified according to either mechanism[4][5] or homology.[6][7]
Contents
Species distribution
Nuclear receptors are specific to metazoans (animals) and are not found in protists, algae, fungi, or plants.[8] Amongst the early-branching animal lineages with sequenced genomes, two have been reported from the sponge Amphimedon queenslandica, two from the ctenophore Mnemiopsis leidyi[9] four from the placozoan Trichoplax adhaerens and 17 from the cnidarian Nematostella vectensis.[10] There are 270 nuclear receptors in the nematode C. elegans alone.[11] Humans, mice, and rats have respectively 48, 49, and 47 nuclear receptors each.[12]
Ligands
Ligands that bind to and activate nuclear receptors include lipophilic substances such as endogenous hormones, vitamins A and D, and xenobiotic endocrine disruptors. Because the expression of a large number of genes is regulated by nuclear receptors, ligands that activate these receptors can have profound effects on the organism. Many of these regulated genes are associated with various diseases, which explains why the molecular targets of approximately 13% of U.S. Food and Drug Administration (FDA) approved drugs are nuclear receptors.[13]
A number of nuclear receptors, referred to as orphan receptors,[14] have no known (or at least generally agreed upon) endogenous ligands. Some of these receptors such as FXR, LXR, and PPAR bind a number of metabolic intermediates such as fatty acids, bile acids and/or sterols with relatively low affinity. These receptors hence may function as metabolic sensors. Other nuclear receptors, such as CAR and PXR appear to function as xenobiotic sensors up-regulating the expression of cytochrome P450 enzymes that metabolize these xenobiotics.[15]
Structure
Nuclear receptors are modular in structure and contain the following domains:[16][17]
- A-B) N-terminal regulatory domain: Contains the activation function 1 (AF-1) whose action is independent of the presence of ligand.[18] The transcriptional activation of AF-1 is normally very weak, but it does synergize with AF-2 in the E-domain (see below) to produce a more robust upregulation of gene expression. The A-B domain is highly variable in sequence between various nuclear receptors.
- C) DNA-binding domain (DBD): Highly conserved domain containing two zinc fingers that binds to specific sequences of DNA called hormone response elements (HRE).
- D) Hinge region: Thought to be a flexible domain that connects the DBD with the LBD. Influences intracellular trafficking and subcellular distribution.
- E) Ligand binding domain (LBD): Moderately conserved in sequence and highly conserved in structure between the various nuclear receptors. The structure of the LBD is referred to as an alpha helical sandwich fold in which three anti parallel alpha helices (the "sandwich filling") are flanked by two alpha helices on one side and three on the other (the "bread"). The ligand binding cavity is within the interior of the LBD and just below three anti parallel alpha helical sandwich "filling". Along with the DBD, the LBD contributes to the dimerization interface of the receptor and in addition, binds coactivator and corepressor proteins. The LBD also contains the activation function 2 (AF-2) whose action is dependent on the presence of bound ligand.[18]
- F) C-terminal domain: Highly variable in sequence between various nuclear receptors.
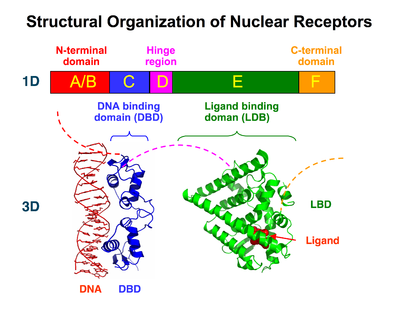 Structural Organization of Nuclear Receptors
Structural Organization of Nuclear Receptors
Top – Schematic 1D amino acid sequence of a nuclear receptor.
Bottom – 3D structures of the DBD (bound to DNA) and LBD (bound to hormone) regions of the nuclear receptor. The structures shown are of the estrogen receptor. Experimental structures of N-terminal domain (A/B), hinge region (D), and C-terminal domain (F) have not been determined therefore are represented by red, purple, and orange dashed lines, respectively.DNA binding domain (DBD) 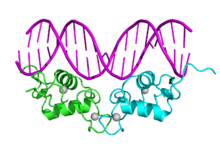
Crystallographic structure of the human progesterone receptor DNA-binding domain dimer (cyan and green) complexed with double strained DNA (magenta). Zinc atoms of are depicted as grey spheres.[19] Identifiers Symbol zf-C4 Pfam PF00105 InterPro IPR001628 SMART SM00399 PROSITE PDOC00031 SCOP 1hra CDD cd06916 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Ligand-binding domain (LBD) 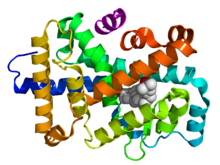
Crystallographic structure of the ligand binding domain of the human RORγ (rainbow colored, N-terminus = blue, C-terminus = red) complexed with 25-hydroxycholesterol (space-filling model (carbon = white, oxygen = red) and the NCOA2 coactivator (magneta).[20] Identifiers Symbol Hormone_recep Pfam PF00104 InterPro IPR000536 SMART SM00430 SCOP 1lbd CDD cd06157 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Mechanism of action
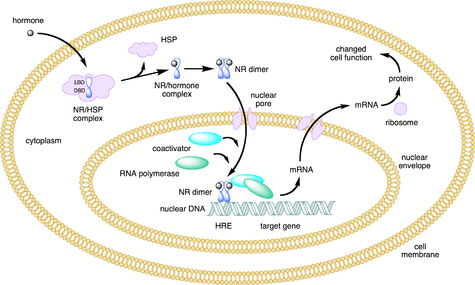 Mechanism nuclear receptor action. This figure depicts the mechanism of a class I nuclear receptor (NR) that, in the absence of ligand, is located in the cytosol. Hormone binding to the NR triggers dissociation of heat shock proteins (HSP), dimerization, and translocation to the nucleus, where the NR binds to a specific sequence of DNA known as a hormone response element (HRE). The nuclear receptor DNA complex in turn recruits other proteins that are responsible for transcription of downstream DNA into mRNA, which is eventually translated into protein, which results in a change in cell function.
Mechanism nuclear receptor action. This figure depicts the mechanism of a class I nuclear receptor (NR) that, in the absence of ligand, is located in the cytosol. Hormone binding to the NR triggers dissociation of heat shock proteins (HSP), dimerization, and translocation to the nucleus, where the NR binds to a specific sequence of DNA known as a hormone response element (HRE). The nuclear receptor DNA complex in turn recruits other proteins that are responsible for transcription of downstream DNA into mRNA, which is eventually translated into protein, which results in a change in cell function.
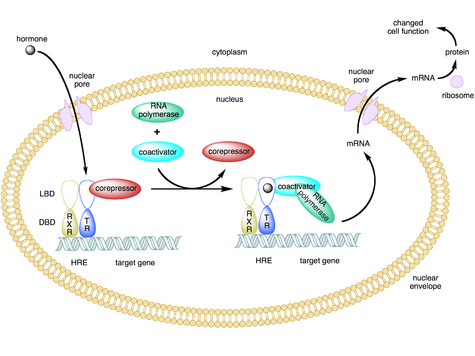 Mechanism nuclear receptor action. This figure depicts the mechanism of a class II nuclear receptor (NR), which, regardless of ligand-binding status, is located in the nucleus bound to DNA. For the purpose of illustration, the nuclear receptor shown here is the thyroid hormone receptor (TR) heterodimerized to the RXR. In the absence of ligand, the TR is bound to corepressor protein. Ligand binding to TR causes a dissociation of corepressor and recruitment of coactivator protein, which, in turn, recruits additional proteins such as RNA polymerase that are responsible for transcription of downstream DNA into RNA and eventually protein, which results in a change in cell function.
Mechanism nuclear receptor action. This figure depicts the mechanism of a class II nuclear receptor (NR), which, regardless of ligand-binding status, is located in the nucleus bound to DNA. For the purpose of illustration, the nuclear receptor shown here is the thyroid hormone receptor (TR) heterodimerized to the RXR. In the absence of ligand, the TR is bound to corepressor protein. Ligand binding to TR causes a dissociation of corepressor and recruitment of coactivator protein, which, in turn, recruits additional proteins such as RNA polymerase that are responsible for transcription of downstream DNA into RNA and eventually protein, which results in a change in cell function.
Nuclear receptors (NRs) may be classified into two broad classes according to their mechanism of action and subcellular distribution in the absence of ligand.
Small lipophilic substances such as natural hormones diffuse past the cell membrane and bind to nuclear receptors located in the cytosol (type I NR) or nucleus (type II NR) of the cell. This causes a change in the conformation of the receptor, which, depending on the mechanistic class (type I or II), triggers a number of down stream events that eventually results in up or down regulation of gene expression. In addition, two additional classes, type III which are a variant of type I, and type IV that bind DNA as monomers have also been defined.[4]
Accordingly, nuclear receptors may be subdivided into the following four mechanistic classes:[4][5]
Type I
Ligand binding to type I nuclear receptors in the cytosol results in the dissociation of heat shock proteins, homo-dimerization, translocation (i.e., active transport) from the cytoplasm into the cell nucleus, and binding to specific sequences of DNA known as hormone response elements (HREs). Type I nuclear receptors bind to HREs consisting of two half-sites separated by a variable length of DNA, and the second half-site has a sequence inverted from the first (inverted repeat). Type I nuclear receptors include members of subfamily 3, such as the androgen receptor, estrogen receptors, glucocorticoid receptor, and progesterone receptor.[21]
It has been noted that some of the NR subfamily 2 nuclear receptors may bind to direct repeat instead of inverted repeat HREs. In addition, some nuclear receptors that bind either as monomers or dimers, with only a single DNA binding domain of the receptor attaching to a single half site HRE. As of now, these nuclear receptors are considered orphan receptors, as their endogenous ligands still unknown.
The nuclear receptor/DNA complex then recruits other proteins that transcribe DNA downstream from the HRE into messenger RNA and eventually protein, which causes a change in cell function.
Type II
Type II receptors, in contrast to type I, are retained in the nucleus regardless of the ligand binding status and in addition bind as hetero-dimers (usually with RXR) to DNA. In the absence of ligand, type II nuclear receptors are often complexed with corepressor proteins. Ligand binding to the nuclear receptor causes dissociation of corepressor and recruitment of coactivator proteins. Additional proteins including RNA polymerase are then recruited to the NR/DNA complex that transcribe DNA into messenger RNA.
Type II nuclear receptors include principally subfamily 1, for example the retinoic acid receptor, retinoid X receptor and thyroid hormone receptor.[22]
Type III
Type III nuclear receptors (principally NR subfamily 2) are similar to type I receptors in that both classes bind to DNA as homodimers. However, type III nuclear receptors, in contrast to type I, bind to direct repeat instead of inverted repeat HREs.
Type IV
Type IV nuclear receptors bind either as monomers or dimers, but only a single DNA binding domain of the receptor binds to a single half site HRE. Examples of type IV receptors are found in most of the NR subfamilies.
Coregulatory proteins
Main article: nuclear receptor coregulatorsNuclear receptors bound to hormone response elements recruit a significant number of other proteins (referred to as transcription coregulators) that facilitate or inhibit the transcription of the associated target gene into mRNA.[23][24] The function of these coregulators are varied and include chromatin remodeling (making the target gene either more or less accessible to transcription) or a bridging function to stabilize the binding of other coregulatory proteins.
Coactivators
Binding of agonist ligands (see section below) to nuclear receptors induces a conformation of the receptor that preferentially binds coactivator proteins. These proteins often have an intrinsic histone acetyltransferase (HAT) activity, which weakens the association of histones to DNA, and therefore promotes gene transcription.
Corepressors
Binding of antagonist ligands to nuclear receptors in contrast induces a conformation of the receptor that preferentially binds corepressor proteins. These proteins, in turn, recruit histone deacetylases (HDACs), which strengthens the association of histones to DNA, and therefore represses gene transcription.
Agonism vs antagonism
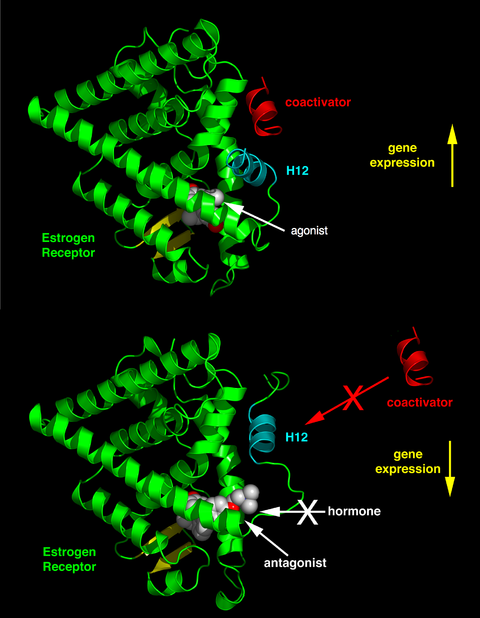 Stuctural basis for the mechanism of nuclear receptor agonist and antagonist action.[25] The structures shown here are of the ligand binding domain (LBD) of the estrogen receptor (green cartoon diagram) complexed with either the agonist diethylstilbestrol (top, PDB 3ERD) or antagonist 4-hydroxytamoxifen (bottom, 3ERT). The ligands are depicted as space filling spheres (white = carbon, red = oxygen). When an agonist is bound to a nuclear receptor, the C-terminal alpha helix of the LDB (H12; light blue) is positioned such that a coactivator protein (red) can bind to the surface of the LBD. Shown here is just a small part of the coactivator protein, the so called NR box containing the LXXLL amino acid sequence motif.[26] Antagonists occupy the same ligand binding cavity of the nuclear receptor. However antagonist ligands in addition have a sidechain extension which sterically displaces H12 to occupy roughly the same position in space as coactivators bind. Hence coactivator binding to the LBD is blocked.
Stuctural basis for the mechanism of nuclear receptor agonist and antagonist action.[25] The structures shown here are of the ligand binding domain (LBD) of the estrogen receptor (green cartoon diagram) complexed with either the agonist diethylstilbestrol (top, PDB 3ERD) or antagonist 4-hydroxytamoxifen (bottom, 3ERT). The ligands are depicted as space filling spheres (white = carbon, red = oxygen). When an agonist is bound to a nuclear receptor, the C-terminal alpha helix of the LDB (H12; light blue) is positioned such that a coactivator protein (red) can bind to the surface of the LBD. Shown here is just a small part of the coactivator protein, the so called NR box containing the LXXLL amino acid sequence motif.[26] Antagonists occupy the same ligand binding cavity of the nuclear receptor. However antagonist ligands in addition have a sidechain extension which sterically displaces H12 to occupy roughly the same position in space as coactivators bind. Hence coactivator binding to the LBD is blocked.
Depending on the receptor involved, the chemical structure of the ligand and the tissue that is being affected, nuclear receptor ligands may display dramatically diverse effects ranging in a spectrum from agonism to antagonism to inverse agonism.[27]
Agonists
The activity of endogenous ligands (such as the hormones estradiol and testosterone) when bound to their cognate nuclear receptors is normally to upregulate gene expression. This stimulation of gene expression by the ligand is referred to as an agonist response. The agonistic effects of endogenous hormones can also be mimicked by certain synthetic ligands, for example, the glucocorticoid receptor anti-inflammatory drug dexamethasone. Agonist ligands work by inducing a conformation of the receptor which favors coactivator binding (see upper half of the figure to the right).
Antagonists
Other synthetic nuclear receptor ligands have no apparent effect on gene transcription in the absence of endogenous ligand. However they block the effect of agonist through competitive binding to the same binding site in the nuclear receptor. These ligands are referred to as antagonists. An example of antagonistic nuclear receptor drug is mifepristone which binds to the glucocorticoid and progesterone receptors and therefore blocks the activity of the endogenous hormones cortisol and progesterone respectively. Antagonist ligands work by inducing a conformation of the receptor which prevents coactivator and promotes corepressor binding (see lower half of the figure to the right).
Inverse agonists
Finally, some nuclear receptors promote a low level of gene transcription in the absence of agonists (also referred to as basal or constitutive activity). Synthetic ligands which reduce this basal level of activity in nuclear receptors are known as inverse agonists.[28]
Selective receptor modulators
A number of drugs that work through nuclear receptors display an agonist response in some tissues and an antagonistic response in other tissues. This behavior may have substantial benefits since it may allow retaining the desired beneficial therapeutic effects of a drug while minimizing undesirable side effects. Drugs with this mixed agonist/antagonist profile of action are referred to as selective receptor modulators (SRMs). Examples include Selective Androgen Receptor Modulators (SARMs), Selective Estrogen Receptor Modulators (SERMs) and Selective Progesterone Receptor Modulators (SPRMs). The mechanism of action of SRMs may vary depending on the chemical structure of the ligand and the receptor involved, however it is thought that many SRMs work by promoting a conformation of the receptor that is closely balanced between agonism and antagonism. In tissues where the concentration of coactivator proteins is higher than corepressors, the equilibrium is shifted in the agonist direction. Conversely in tissues where corepressors dominate, the ligand behaves as an antagonist.[29]
Alternative mechanisms
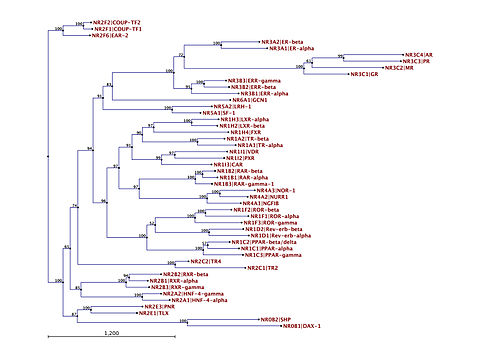 Phylogenetic tree of human nuclear receptors
Phylogenetic tree of human nuclear receptors
Transrepression
The most common mechanism of nuclear receptor action involves direct binding of the nuclear receptor to a DNA hormone response element. This mechanism is referred to as transactivation. However some nuclear receptors not only have the ability to directly bind to DNA, but also to other transcription factors. This binding often results in deactivation of the second transcription factor in a process known as transrepression.[30] One example of a nuclear receptor that are able to transrepress is the glucocorticoid receptor (GR). Furthermore certain GR ligands known as Selective Glucocorticoid Receptor Agonists (SEGRAs) are able to activate GR in such a way that GR more strongly transrepresses than transactivates. This selectivity increases the separation between the desired antiinflammatory effects and undesired metabolic side effects of these selective glucocorticoids.
Non-genomic
The classical direct effects of nuclear receptors on gene regulation normally takes hours before a functional effect is seen in cells because of the large number of intermediate steps between nuclear receptor activation and changes in protein expression levels. However it has been observed that some effects from the application of hormones such as estrogen occur within minutes which is inconsistent with the classical mechanism of nuclear receptor action. While the molecular target for these non-genomic effects of nuclear receptors has not been conclusively demonstrated, it has been hypothesized that there are variants of nuclear receptors which are membrane associated instead of being localized in the cytosol or nucleus. Furthermore these membrane associated receptors function through alternative signal transduction mechanisms not involving gene regulation.[31][32]
Family members
The following is a list of the 48 known human nuclear receptors[12] categorized according to sequence homology.[6][7]
Subfamily Group Member NRNC Symbol[6] Abbreviation Name Gene Ligand(s) 1 Thyroid Hormone Receptor-like A Thyroid hormone receptor NR1A1 TRα Thyroid hormone receptor-α THRA thyroid hormone NR1A2 TRβ Thyroid hormone receptor-β THRB B Retinoic acid receptor NR1B1 RARα Retinoic acid receptor-α RARA vitamin A and related compounds NR1B2 RARβ Retinoic acid receptor-β RARB NR1B3 RARγ Retinoic acid receptor-γ RARG C Peroxisome proliferator-activated receptor NR1C1 PPARα Peroxisome proliferator-activated receptor-α PPARA fatty acids, prostaglandins NR1C2 PPAR-β/δ Peroxisome proliferator-activated receptor-β/δ PPARD NR1C3 PPARγ Peroxisome proliferator-activated receptor-γ PPARG D Rev-ErbA NR1D1 Rev-ErbAα Rev-ErbAα NR1D1 heme NR1D2 Rev-ErbAβ Rev-ErbAα NR1D2 F RAR-related orphan receptor NR1F1 RORα RAR-related orphan receptor-α RORA cholesterol, ATRA NR1F2 RORβ RAR-related orphan receptor-β RORB NR1F3 RORγ RAR-related orphan receptor-γ RORC H Liver X receptor-like NR1H3 LXRα Liver X receptor-α NR1H3 oxysterols NR1H2 LXRβ Liver X receptor-β NR1H2 NR1H4 FXR Farnesoid X receptor NR1H4 I Vitamin D receptor-like NR1I1 VDR Vitamin D receptor VDR vitamin D NR1I2 PXR Pregnane X receptor NR1I2 xenobiotics NR1I3 CAR Constitutive androstane receptor NR1I3 androstane X NRs with two DNA binding domains[33][34] NR1X1 2DBD-NRα NR1X2 2DBD-NRβ NR1X3 2DBD-NRγ 2 Retinoid X Receptor-like A Hepatocyte nuclear factor-4 NR2A1 HNF4α Hepatocyte nuclear factor-4-α HNF4A fatty acids NR2A2 HNF4γ Hepatocyte nuclear factor-4-γ HNF4G B Retinoid X receptor NR2B1 RXRα Retinoid X receptor-α RXRA retinoids NR2B2 RXRβ Retinoid X receptor-β RXRB NR2B3 RXRγ Retinoid X receptor-γ RXRG C Testicular receptor NR2C1 TR2 Testicular receptor 2 NR2C1 NR2C2 TR4 Testicular receptor 4 NR2C2 E TLX/PNR NR2E1 TLX Homologue of the Drosophila tailless gene NR2E1 NR2E3 PNR Photoreceptor cell-specific nuclear receptor NR2E3 F COUP/EAR NR2F1 COUP-TFI Chicken ovalbumin upstream promoter-transcription factor I NR2E1 NR2F2 COUP-TFII Chicken ovalbumin upstream promoter-transcription factor II NR2F2 NR2F6 EAR-2 V-erbA-related NR2F6 3 Estrogen Receptor-like A Estrogen receptor NR3A1 ERα Estrogen receptor-α ESR1 estrogens NR3A2 ERβ Estrogen receptor-β ESR2 B Estrogen related receptor NR3B1 ERRα Estrogen-related receptor-α ESRRA NR3B2 ERRβ Estrogen-related receptor-β ESRRB NR3B3 ERRγ Estrogen-related receptor-γ ESRRG C 3-Ketosteroid receptors NR3C1 GR Glucocorticoid receptor NR3C1 cortisol NR3C2 MR Mineralocorticoid receptor NR3C2 aldosterone NR3C3 PR Progesterone receptor PGR progesterone NR3C4 AR Androgen receptor AR testosterone 4 Nerve Growth Factor IB-like A NGFIB/NURR1/NOR1 NR4A1 NGFIB Nerve Growth factor IB NR4A1 NR4A2 NURR1 Nuclear receptor related 1 NR4A2 NR4A3 NOR1 Neuron-derived orphan receptor 1 NR4A3 5 Steroidogenic Factor-like A SF1/LRH1 NR5A1 SF1 Steroidogenic factor 1 NR5A1 phosphatidylinositols NR5A2 LRH-1 Liver receptor homolog-1 NR5A2 phosphatidylinositols 6 Germ Cell Nuclear Factor-like A GCNF NR6A1 GCNF Germ cell nuclear factor NR6A1 0 Miscellaneous A DAX/SHP NR0B1 DAX1 Dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 NR0B1 NR0B2 SHP Small heterodimer partner NR0B2 History
Below is a brief selection of key events in the history of nuclear receptor research.[35]
- 1905 – Ernest Starling coined the word hormone
- 1926 – Edward Calvin Kendall and Tadeus Reichstein isolated and determined the structures of cortisone and thyroxine
- 1929 – Adolf Butenandt and Edward Adelbert Doisy – independently isolated and determined the structure of estrogen
- 1958 – Elwood Jensen – isolated the estrogen receptor
- 1980s – cloning of the estrogen, glucocorticoid, and thyroid hormone receptors by Pierre Chambon, Ronald Evans, and Björn Vennström respectively
- 2004 – Pierre Chambon, Ronald Evans, and Elwood Jensen were awarded the Albert Lasker Award for Basic Medical Research, an award that frequently precedes a Nobel Prize in Medicine
See also
References
- ^ PDB 3E00; Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature. 2008:350–356. doi:10.1038/nature07413. PMID 18971932.
- ^ Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240(4854):889–95. doi:10.1126/science.3283939. PMID 3283939.
- ^ Olefsky JM. Nuclear receptor minireview series. J. Biol. Chem.. 2001;276(40):36863–4. doi:10.1074/jbc.R100047200. PMID 11459855.
- ^ a b c Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–9. doi:10.1016/0092-8674(95)90199-X. PMID 8521507.
- ^ a b Novac N, Heinzel T. Nuclear receptors: overview and classification. Curr Drug Targets Inflamm Allergy. 2004;3(4):335–46. doi:10.2174/1568010042634541. PMID 15584884.
- ^ a b c Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97(2):161–3. doi:10.1016/S0092-8674(00)80726-6. PMID 10219237.
- ^ a b Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J. Mol. Endocrinol.. 1997;19(3):207–26. doi:10.1677/jme.0.0190207. PMID 9460643.
- ^ Escriva H, Langlois MC, Mendonça RL, Pierce R, Laudet V. Evolution and diversification of the nuclear receptor superfamily. Annals of the New York Academy of Sciences. 1998;839:143–6. doi:10.1111/j.1749-6632.1998.tb10747.x. PMID 9629140.
- ^ Reitzel AM, Pang K, Ryan JF, Mullikin JC, Martindale MQ, Baxevanis AD, Tarrant AM. Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily?. Evodevo. 2011;2(1):3. doi:10.1186/2041-9139-2-3. PMID 21291545.
- ^ Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier ME, Ortlund EA, Degnan BM, Thornton JW. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol.. 2010;8(10). doi:10.1371/journal.pbio.1000497. PMID 20957188.
- ^ Sluder AE, Maina CV. Nuclear receptors in nematodes: themes and variations. Trends in Genetics : TIG. 2001;17(4):206–13. doi:10.1016/S0168-9525(01)02242-9. PMID 11275326.
- ^ a b Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14(4):580–90. doi:10.1101/gr.2160004. PMID 15059999.
- ^ Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there?. Nature reviews. Drug discovery. 2006;5(12):993–6. doi:10.1038/nrd2199. PMID 17139284.
- ^ Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, Perlmann T, Renaud JP, Schwabe J, Sladek F, Tsai MJ, Laudet V. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol. Rev.. 2006;58(4):798–836. doi:10.1124/pr.58.4.10. PMID 17132856.
- ^ Mohan R, Heyman RA. Orphan nuclear receptor modulators. Curr Top Med Chem. 2003;3(14):1637–47. doi:10.2174/1568026033451709. PMID 14683519.
- ^ Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64(5):310–9. doi:10.1016/S0039-128X(99)00014-8. PMID 10406480.
- ^ Klinge CM. Estrogen receptor interaction with co-activators and co-repressors. Steroids. 2000;65(5):227–51. doi:10.1016/S0039-128X(99)00107-5. PMID 10751636.
- ^ a b Wärnmark A, Treuter E, Wright AP, Gustafsson J-Å. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol. Endocrinol.. 2003;17(10):1901–9. doi:10.1210/me.2002-0384. PMID 12893880.
- ^ PDB 2C7A; Roemer SC, Donham DC, Sherman L, Pon VH, Edwards DP, Churchill ME. Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements. Mol. Endocrinol.. 2006;20(12):3042–52. doi:10.1210/me.2005-0511. PMID 16931575.
- ^ PDB 3L0L; Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol. Endocrinol.. 2010;24(5):923–9. doi:10.1210/me.2009-0507. PMID 20203100.
- ^ Linja MJ, Porkka KP, Kang Z, et al.. Expression of androgen receptor coregulators in prostate cancer. Clin. Cancer Res.. 2004;10(3):1032–40. doi:10.1158/1078-0432.CCR-0990-3. PMID 14871982.
- ^ Klinge CM, Bodenner DL, Desai D, Niles RM, Traish AM. Binding of type II nuclear receptors and estrogen receptor to full and half-site estrogen response elements in vitro. Nucleic Acids Res.. 1997;25(10):1903–12. doi:10.1093/nar/25.10.1903. PMID 9115356.
- ^ Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14(2):121–41. doi:10.1101/gad.14.2.121. PMID 10652267.
- ^ Aranda A, Pascual A. Nuclear hormone receptors and gene expression [abstract]. Physiol. Rev.. 2001;81(3):1269–304. PMID 11427696.
- ^ Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, Öhman L, Greene GL, Gustafsson J-Å, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389(6652):753–8. doi:10.1038/39645. PMID 9338790.
- ^ Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–37. doi:10.1016/S0092-8674(00)81717-1. PMID 9875847.
- ^ Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nature reviews. Drug discovery. 2004;3(11):950–64. doi:10.1038/nrd1551. PMID 15520817.
- ^ Busch BB, Stevens WC, Martin R, et al.. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J. Med. Chem.. 2004;47(23):5593–6. doi:10.1021/jm049334f. PMID 15509154.
- ^ Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. doi:10.1210/er.2003-0023. PMID 14769827.
- ^ Pascual G, Glass CK. Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol Metab. 2006;17(8):321–7. doi:10.1016/j.tem.2006.08.005. PMID 16942889.
- ^ Björnström L, Sjöberg M. Estrogen receptor-dependent activation of AP-1 via non-genomic signalling. Nucl Recept. 2004;2(1):3. doi:10.1186/1478-1336-2-3. PMID 15196329.
- ^ Zivadinovic D, Gametchu B, Watson CS. Membrane estrogen receptor-alpha levels in MCF-7 breast cancer cells predict cAMP and proliferation responses. Breast Cancer Res.. 2005;7(1):R101–12. doi:10.1186/bcr958. PMID 15642158.
- ^ Wu W, Niles EG, El-Sayed N, Berriman M, LoVerde PT. Schistosoma mansoni (Platyhelminthes, Trematoda) nuclear receptors: sixteen new members and a novel subfamily. Gene. 2006;366(2):303–15. doi:10.1016/j.gene.2005.09.013. PMID 16406405.
- ^ Wu W, Niles EG, Hirai H, LoVerde PT. Evolution of a novel subfamily of nuclear receptors with members that each contain two DNA binding domains. BMC Evol Biol. 2007;7(Feb 23):27. doi:10.1186/1471-2148-7-27. PMID 17319953.
- ^ Tata JR. One hundred years of hormones. EMBO Rep.. 2005;6(6):490–6. doi:10.1038/sj.embor.7400444. PMID 15940278.
External links
- Eukaryotic Linear Motif resource motif class LIG_CORNRBOX
- Eukaryotic Linear Motif resource motif class LIG_NRBOX
- MeSH Nuclear+Receptors
- Vincent Laudet. The International Union of Basic and Clinical Pharmacology. The IUPHAR Compendium of the Pharmacology and Classification of the Nuclear Receptor Superfamily 2006E; 2006 [cited 2008-02-21].
- published by BioMed Central (no longer accepting submissions since May 2007). Nuclear Receptor online journal [cited 2008-02-21].
- Georgetown University. Nuclear Receptor Resource [cited 2008-02-21].
- The NURSA Consortium. Nuclear Receptor Signaling Atlas (Receptors, Coactivators, Corepressors and Ligands) [cited 2008-02-21]. "an NIH-funded research consortium and database; includes open-access PubMed-indexed journal, Nuclear Receptor Signaling"
- Jack Vanden Heuvel. Nuclear Receptor Resource [cited 2009-09-21].
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) Miscellaneoussee also transcription factor/coregulator deficiencies
B bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°)Categories:- Intracellular receptors
- Protein families
- Transcription factors
Wikimedia Foundation. 2010.