- Microphthalmia-associated transcription factor
-
Microphthalmia-associated transcription factor Identifiers Symbols MITF; MI; WS2; WS2A; bHLHe32 External IDs OMIM: 156845 MGI: 104554 HomoloGene: 4892 GeneCards: MITF Gene Gene Ontology Molecular function • DNA binding Cellular component • nucleus Biological process • regulation of transcription, DNA-dependent
• multicellular organismal development
• melanocyte differentiation
• positive regulation of transcription, DNA-dependent
• positive regulation of transcription, DNA-dependentSources: Amigo / QuickGO RNA expression pattern 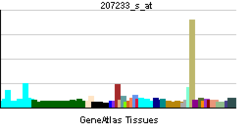
More reference expression data Orthologs Species Human Mouse Entrez 4286 17342 Ensembl ENSG00000187098 ENSMUSG00000035158 UniProt O75030 Q3U2D2 RefSeq (mRNA) NM_000248.3 NM_008601 RefSeq (protein) NP_000239.1 NP_032627 Location (UCSC) Chr 3:
69.79 – 70.02 MbChr 6:
97.76 – 97.97 MbPubMed search [1] [2] Microphthalmia-associated transcription factor (Mitf) is a basic helix-loop-helix leucine zipper transcription factor involved in melanocyte[1] and osteoclast development.[2]
Contents
Clinical significance
It can be associated with Tietz syndrome[3] and Waardenburg syndrome type IIa.[4]
Target genes
Mitf recognizes E-box (CAYRTG) and M-box (TCAYRTG or CAYRTGA) sequences in the promoter regions of target genes. Known target genes (confirmed by at least two independent sources) of this transcription factor include,
Additional genes identified by a microarray study (which confirmed the above targets) include the following,[6]
MBP TNFRSF14 IRF4 RBM35A PLA1A APOLD1 KCNN2 INPP4B CAPN3 LGALS3 GREB1 FRMD4B SLC1A4 TBC1D16 GMPR ASAH1 MICAL1 TMC6 ITPKB SLC7A8 Interactions
Microphthalmia-associated transcription factor has been shown to interact with PATZ1,[24] PIAS3,[25] TFE3,[26][27] UBE2I[28] and Lymphoid enhancer-binding factor 1.[12]
See also
References
- ^ Levy C, Khaled M, Fisher DE (2006). "MITF: master regulator of melanocyte development and melanoma oncogene". Trends Mol Med 12 (9): 406–14. doi:10.1016/j.molmed.2006.07.008. PMID 16899407.
- ^ Hershey CL, Fisher DE (2004). "Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function". Bone 34 (4): 689–96. doi:10.1016/j.bone.2003.08.014. PMID 15050900.
- ^ Smith SD, Kelley PM, Kenyon JB, Hoover D (2000). "Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF". J. Med. Genet. 37 (6): 446–8. doi:10.1136/jmg.37.6.446. PMC 1734605. PMID 10851256. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1734605.
- ^ Tachibana M, Kobayashi Y, Matsushima Y (2003). "Mouse models for four types of Waardenburg syndrome". Pigment Cell Res. 16 (5): 448–54. doi:10.1034/j.1600-0749.2003.00066.x. PMID 12950719.
- ^ Luchin A, Purdom G, Murphy K, et al. (2000). "The microphthalmia transcription factor reulates expression of the tartrate-resistant acid phosphatase gene during terminal differentiation of osteoclasts". J. Bone Miner. Res. 15 (3): 451–460. doi:10.1359/jbmr.2000.15.3.451. PMID 10750559.
- ^ a b c d e f g h i j k l m n o p q r s t u Hoek KS, Schlegel NC, Eichhoff OM, et al. (2008). "Novel MITF targets identified using a two-step DNA microarray strategy". Pigment Cell Melanoma Res. 21 (6): 665–76. doi:10.1111/j.1755-148X.2008.00505.x. PMID 19067971.
- ^ McGill GG, Horstmann M, Widlund HR, et al. (2002). "Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability". Cell 109 (6): 707–18. doi:10.1016/S0092-8674(02)00762-6. PMID 12086670.
- ^ Esumi N, Kachi S, Campochiaro PA, et al. (2007). "VMD2 promoter requires two proximal E-box sites for its activity in vivo and is regulated by the MITF-TFE family". J. Biol. Chem. 282 (3): 1838–50. doi:10.1074/jbc.M609517200. PMID 17085443.
- ^ Dynek JN, Chan SM, Liu J, et al. (2008). "Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas". Cancer Res. 68 (9): 3124–32. doi:10.1158/0008-5472.CAN-07-6622. PMID 18451137.
- ^ Du J, Widlund HR, Horstmann MA, et al. (2004). "Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF". Cancer Cell 6 (6): 565–76. doi:10.1016/j.ccr.2004.10.014. PMID 15607961.
- ^ a b Meadows NA, Sharma SM, Faulkner GJ, et al. (2007). "The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor". J. Biol. Chem. 282 (3): 1891–904. doi:10.1074/jbc.M608572200. PMID 17105730.
- ^ a b Yasumoto K, Takeda K, Saito H, et al. (2002). "Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling". EMBO J. 21 (11): 2703–14. doi:10.1093/emboj/21.11.2703. PMC 126018. PMID 12032083. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=126018.
- ^ Sato-Jin K, Nishimura EK, Akasaka E, et al. (2008). "Epistatic connections between microphthalmia-associated transcription factor and endothelin signaling in Waardenburg syndrome and other pigmentary disorders". FASEB J. 22 (4): 1155–68. doi:10.1096/fj.07-9080com. PMID 18039926.
- ^ Loftus SK, Antonellis A, Matera I, et al. (2009). "Gpnmb is a Melanoblast-Expressed, MITF-Dependent Gene". Pigment Cell Melanoma Res. 22 (1): 99–110. doi:10.1111/j.1755-148X.2008.00518.x. PMC 2714741. PMID 18983539. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2714741.
- ^ Vetrini F, Auricchio A, Du J, Angeletti B, et al. (2004). "The microphthalmia transcription factor (Mitf) controls expression of the ocular albinism type 1 gene: link between melanin synthesis and melanosome biogenesis". Mol. Cell. Biol. 24 (15): 6550–9. doi:10.1128/MCB.24.15.6550-6559.2004. PMC 444869. PMID 15254223. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=444869.
- ^ Aoki H, Moro O (2002). "Involvement of microphthalmia-associated transcription factor (MITF) in expression of human melanocortin-1 receptor (MC1R)". Life Sci. 71 (18): 2171–9. doi:10.1016/S0024-3205(02)01996-3. PMID 12204775.
- ^ a b Du J, Miller AJ, Widlund HR, et al. (2003). "MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma". Am. J. Pathol. 163 (1): 333–43. doi:10.1016/S0002-9440(10)63657-7. PMC 1868174. PMID 12819038. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1868174.
- ^ Chiaverini C, Beuret L, Flori E, et al. (2008). "Microphthalmia-associated transcription factor regulates RAB27A gene expression and controls melanosome transport". J. Biol. Chem. 283 (18): 12635–42. doi:10.1074/jbc.M800130200. PMID 18281284.
- ^ Du J, Fisher DE (2002). "Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF". J. Biol. Chem. 277 (1): 402–6. doi:10.1074/jbc.M110229200. PMID 11700328.
- ^ Carreira S, Liu B, Goding CR (2000). "The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes". J. Biol. Chem. 275 (29): 21920–7. doi:10.1074/jbc.M000035200. PMID 10770922.
- ^ Miller AJ, Du J, Rowan S, et al. (2004). "Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma". Cancer Res. 64 (2): 509–16. doi:10.1158/0008-5472.CAN-03-2440. PMID 14744763.
- ^ Hou L, Panthier JJ, Arnheiter H (2000). "Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF". Development 127 (24): 5379–89. PMID 11076759.
- ^ Fang D, Tsuji Y, Setaluri V (2002). "Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF". Nucleic Acids Res. 30 (14): 3096–106. doi:10.1093/nar/gkf424. PMC 135745. PMID 12136092. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=135745.
- ^ Morii, Eiichi; Oboki Keisuke, Kataoka Tatsuki R, Igarashi Kazuhiko, Kitamura Yukihiko (Mar. 2002). "Interaction and cooperation of mi transcription factor (MITF) and myc-associated zinc-finger protein-related factor (MAZR) for transcription of mouse mast cell protease 6 gene". J. Biol. Chem. (United States) 277 (10): 8566–71. doi:10.1074/jbc.M110392200. ISSN 0021-9258. PMID 11751862.
- ^ Levy, Carmit; Nechushtan Hovav, Razin Ehud (Jan. 2002). "A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor". J. Biol. Chem. (United States) 277 (3): 1962–6. doi:10.1074/jbc.M109236200. ISSN 0021-9258. PMID 11709556.
- ^ Steingrimsson, Eiríkur; Tessarollo Lino, Pathak Bhavani, Hou Ling, Arnheiter Heinz, Copeland Neal G, Jenkins Nancy A (Apr. 2002). "Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development". Proc. Natl. Acad. Sci. U.S.A. (United States) 99 (7): 4477–82. doi:10.1073/pnas.072071099. ISSN 0027-8424. PMC 123673. PMID 11930005. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=123673.
- ^ Mansky, Kim C; Sulzbacher Sabine, Purdom Georgia, Nelsen Lori, Hume David A, Rehli Michael, Ostrowski Michael C (Feb. 2002). "The microphthalmia transcription factor and the related helix-loop-helix zipper factors TFE-3 and TFE-C collaborate to activate the tartrate-resistant acid phosphatase promoter". J. Leukoc. Biol. (United States) 71 (2): 304–10. ISSN 0741-5400. PMID 11818452.
- ^ Xu, W; Gong L, Haddad M M, Bischof O, Campisi J, Yeh E T, Medrano E E (Mar. 2000). "Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9". Exp. Cell Res. (UNITED STATES) 255 (2): 135–43. doi:10.1006/excr.2000.4803. ISSN 0014-4827. PMID 10694430.
External links
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) Miscellaneoussee also transcription factor/coregulator deficiencies
B bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°)Categories:- Human proteins
- Transcription factors
- Gene expression
Wikimedia Foundation. 2010.
