- N-Myc
-
V-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) Identifiers Symbols MYCN; MODED; N-myc; NMYC; ODED; bHLHe37 External IDs OMIM: 164840 MGI: 97357 HomoloGene: 3922 GeneCards: MYCN Gene Gene Ontology Molecular function • DNA binding
• sequence-specific DNA binding transcription factor activity
• protein bindingCellular component • chromatin
• nucleusBiological process • transcription, DNA-dependent
• regulation of transcription from RNA polymerase II promoterSources: Amigo / QuickGO RNA expression pattern 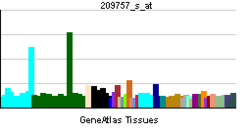
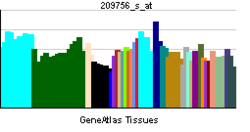
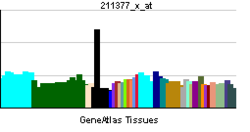
More reference expression data Orthologs Species Human Mouse Entrez 4613 18109 Ensembl ENSG00000134323 ENSMUSG00000037169 UniProt P04198 Q3UII1 RefSeq (mRNA) NM_005378.4 NM_008709.3 RefSeq (protein) NP_005369.2 NP_032735.3 Location (UCSC) Chr 2:
16.08 – 16.09 MbChr 12:
12.94 – 12.95 MbPubMed search [1] [2] V-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian), also known as MYCN, is a protein which in humans is encoded by the MYCN gene.
Contents
Function
This gene is a member of the MYC family of transcription factors and encodes a protein with a basic helix-loop-helix (bHLH) domain. This protein is located in the nucleus and must dimerize with another bHLH protein in order to bind DNA.[1]
Clinical significance
Amplification of this gene is associated with a variety of tumors, most notably neuroblastomas.[2]
Interactions
N-Myc has been shown to interact with MAX.[3][4]
N-Myc is also stabilized by Aurora A which protects it from degradation.[5]
See also
References
- ^ "Entrez Gene: MYCN v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4613.
- ^ Cheng JM, Hiemstra JL, Schneider SS, Naumova A, Cheung NK, Cohn SL, Diller L, Sapienza C, Brodeur GM (June 1993). "Preferential amplification of the paternal allele of the N-myc gene in human neuroblastomas". Nat. Genet. 4 (2): 191–4. doi:10.1038/ng0693-191. PMID 8102299.
- ^ Blackwood, E M; Eisenman R N (Mar. 1991). "Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc". Science (UNITED STATES) 251 (4998): 1211–7. doi:10.1126/science.2006410. ISSN 0036-8075. PMID 2006410.
- ^ FitzGerald, M J; Arsura M, Bellas R E, Yang W, Wu M, Chin L, Mann K K, DePinho R A, Sonenshein G E (Apr. 1999). "Differential effects of the widely expressed dMax splice variant of Max on E-box vs initiator element-mediated regulation by c-Myc". Oncogene (ENGLAND) 18 (15): 2489–98. doi:10.1038/sj.onc.1202611. ISSN 0950-9232. PMID 10229200.
- ^ Stabilization of N-Myc Is a Critical Function of Aurora A in Human Neuroblastoma Cancer Cell, Volume 15, Issue 1, 6 January 2009, Pages 67-78, Tobias Otto, Sebastian Horn, Markus Brockmann, Ursula Eilers, Lars Schüttrumpf, Nikita Popov, Anna Marie Kenney, Johannes H. Schulte, Roderick Beijersbergen, Holger Christiansen, Bernd Berwanger, Martin Eilers
Further reading
- Lüscher B (2001). "Function and regulation of the transcription factors of the Myc/Max/Mad network.". Gene 277 (1-2): 1–14. doi:10.1016/S0378-1119(01)00697-7. PMID 11602341.
- Armstrong BC, Krystal GW (1992). "Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc.". Cell Growth Differ. 3 (6): 385–90. PMID 1419902.
- Hagiwara T, Nakaya K, Nakamura Y, et al. (1992). "Specific phosphorylation of the acidic central region of the N-myc protein by casein kinase II.". Eur. J. Biochem. 209 (3): 945–50. doi:10.1111/j.1432-1033.1992.tb17367.x. PMID 1425701.
- Fougerousse F, Meloni R, Roudaut C, Beckmann JS (1992). "Dinucleotide repeat polymorphism at the human hemoglobin alpha-1 pseudo-gene (HBAP1).". Nucleic Acids Res. 20 (5): 1165. doi:10.1093/nar/20.5.1165. PMC 312136. PMID 1549498. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=312136.
- Krystal GW, Armstrong BC, Battey JF (1990). "N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts.". Mol. Cell. Biol. 10 (8): 4180–91. PMC 360949. PMID 1695323. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=360949.
- Blackwood EM, Eisenman RN (1991). "Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc.". Science 251 (4998): 1211–7. doi:10.1126/science.2006410. PMID 2006410.
- Emanuel BS, Balaban G, Boyd JP, et al. (1985). "N-myc amplification in multiple homogeneously staining regions in two human neuroblastomas.". Proc. Natl. Acad. Sci. U.S.A. 82 (11): 3736–40. doi:10.1073/pnas.82.11.3736. PMC 397862. PMID 2582423. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=397862.
- Ibson JM, Rabbitts PH (1988). "Sequence of a germ-line N-myc gene and amplification as a mechanism of activation.". Oncogene 2 (4): 399–402. PMID 2834684.
- Stanton LW, Schwab M, Bishop JM (1986). "Nucleotide sequence of the human N-myc gene.". Proc. Natl. Acad. Sci. U.S.A. 83 (6): 1772–6. doi:10.1073/pnas.83.6.1772. PMC 323166. PMID 2869488. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=323166.
- Michitsch RW, Melera PW (1985). "Nucleotide sequence of the 3' exon of the human N-myc gene.". Nucleic Acids Res. 13 (7): 2545–58. doi:10.1093/nar/13.7.2545. PMC 341174. PMID 2987858. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=341174.
- Slamon DJ, Boone TC, Seeger RC, et al. (1986). "Identification and characterization of the protein encoded by the human N-myc oncogene.". Science 232 (4751): 768–72. doi:10.1126/science.3008339. PMID 3008339.
- Garson JA, van den Berghe JA, Kemshead JT (1987). "Novel non-isotopic in situ hybridization technique detects small (1 Kb) unique sequences in routinely G-banded human chromosomes: fine mapping of N-myc and beta-NGF genes.". Nucleic Acids Res. 15 (12): 4761–70. doi:10.1093/nar/15.12.4761. PMC 305916. PMID 3299258. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=305916.
- Stanton LW, Bishop JM (1988). "Alternative processing of RNA transcribed from NMYC.". Mol. Cell. Biol. 7 (12): 4266–72. PMC 368108. PMID 3437890. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=368108.
- Kohl NE, Legouy E, DePinho RA, et al. (1986). "Human N-myc is closely related in organization and nucleotide sequence to c-myc.". Nature 319 (6048): 73–7. doi:10.1038/319073a0. PMID 3510398.
- Grady EF, Schwab M, Rosenau W (1987). "Expression of N-myc and c-src during the development of fetal human brain.". Cancer Res. 47 (11): 2931–6. PMID 3552210.
- Ramsay G, Stanton L, Schwab M, Bishop JM (1987). "Human proto-oncogene N-myc encodes nuclear proteins that bind DNA.". Mol. Cell. Biol. 6 (12): 4450–7. PMC 367228. PMID 3796607. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=367228.
- Brodeur GM, Seeger RC (1986). "Gene amplification in human neuroblastomas: basic mechanisms and clinical implications.". Cancer Genet. Cytogenet. 19 (1-2): 101–11. doi:10.1016/0165-4608(86)90377-8. PMID 3940169.
- Kanda N, Schreck R, Alt F, et al. (1983). "Isolation of amplified DNA sequences from IMR-32 human neuroblastoma cells: facilitation by fluorescence-activated flow sorting of metaphase chromosomes.". Proc. Natl. Acad. Sci. U.S.A. 80 (13): 4069–73. doi:10.1073/pnas.80.13.4069. PMC 394202. PMID 6575396. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=394202.
- Schwab M, Varmus HE, Bishop JM, et al. (1984). "Chromosome localization in normal human cells and neuroblastomas of a gene related to c-myc.". Nature 308 (5956): 288–91. doi:10.1038/308288a0. PMID 6700732.
- Brodeur GM, Seeger RC, Schwab M, et al. (1984). "Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage.". Science 224 (4653): 1121–4. doi:10.1126/science.6719137. PMID 6719137.
External links
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) Miscellaneoussee also transcription factor/coregulator deficiencies
B bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°)Categories:- Human proteins
- Transcription factors
- Chromosome 2 gene stubs
Wikimedia Foundation. 2010.
