- NRF1
-
Not to be confused with nuclear factor (erythroid-derived 2)-like 1 or NFE2L1.
Nuclear respiratory factor 1 Identifiers Symbols NRF1; ALPHA-PAL External IDs OMIM: 600879 MGI: 1332235 HomoloGene: 3674 GeneCards: NRF1 Gene Gene Ontology Molecular function • DNA binding Cellular component • nucleus
• cytoplasmBiological process • response to hypoxia
• generation of precursor metabolites and energy
• regulation of transcription from RNA polymerase II promoter
• mitochondrion organization
• organ regeneration
• response to estradiol stimulus
• response to lipopolysaccharide
• response to folic acid
• response to electrical stimulusSources: Amigo / QuickGO RNA expression pattern 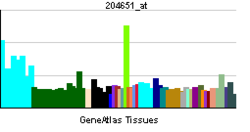
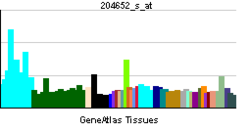
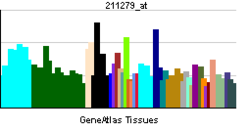
More reference expression data Orthologs Species Human Mouse Entrez 4899 18181 Ensembl ENSG00000106459 ENSMUSG00000058440 UniProt Q16656 Q8BSP8 RefSeq (mRNA) NM_001040110.1 NM_010938 RefSeq (protein) NP_001035199.1 NP_035068 Location (UCSC) Chr 7:
129.25 – 129.4 MbChr 6:
30 – 30.1 MbPubMed search [1] [2] Nuclear respiratory factor 1, also known as Nrf1, Nrf-1, NRF1 and NRF-1, encodes a protein that homodimerizes and functions as a transcription factor which activates the expression of some key metabolic genes regulating cellular growth and nuclear genes required for respiration, heme biosynthesis, and mitochondrial DNA transcription and replication. The protein has also been associated with the regulation of neurite outgrowth. Alternate transcriptional splice variants, which encode the same protein, have been characterized. Additional variants encoding different protein isoforms have been described but they have not been fully characterized. Confusion has occurred in bibliographic databases due to the shared symbol of NRF1 for this gene and for "nuclear factor (erythroid-derived 2)-like 1" which has an official symbol of NFE2L1. [provided by RefSeq]
Contents
Function
Nrf1 functions as a transcription factor that activates the expression of some key metabolic genes regulating cellular growth and nuclear genes required for mitochondrial respiration, and mitochondrial DNA transcription and replication. Nrf1, together with Nrf2, mediates the biogenomic coordination between nuclear and mitochondrial genomes by directly regulating the expression of several nuclear-encoded ETC proteins, and indirectly regulating the three mitochondrial-encoded COX subunit genes by activating mtTFA, mtTFB1, and mtTFB2. The Nrf proteins are also important for the upregulation of antioxidant and xenobiotic-metabolizing enzymes during oxidative stress.[1]
Nrf1 is also involved in heme biosynthesis through regulation of globin gene expression.[2] The protein has also been associated with the regulation of neurite outgrowth.[3]
Alternate transcriptional splice variants, which encode the same protein, have been characterized. Additional variants encoding different protein isoforms have been described but they have not been fully characterized.[4]
Interactions
Nrf1 heterodimerizes with small Maf proteins and the resultant heterodimers bind to the antioxidant response element (ARE) thereby mediating the transcription of antioxidative and phase 2-metabolizing enzymes.[1][5]
NRF1 has been shown to interact with DYNLL1[6] and PPARGC1A.[7]
References
- ^ a b Biswas M, Chan JY (April 2010). "Role of Nrf1 in antioxidant response element-mediated gene expression and beyond". Toxicol. Appl. Pharmacol. 244 (1): 16–20. doi:10.1016/j.taap.2009.07.034. PMC 2837788. PMID 19665035. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2837788.
- ^ Chan JY, Han XL, Kan YW (December 1993). "Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast". Proc. Natl. Acad. Sci. U.S.A. 90 (23): 11371–5. doi:10.1073/pnas.90.23.11371. PMC 47984. PMID 8248256. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=47984.
- ^ Chang WT, Chen HI, Chiou RJ, Chen CY, Huang AM (August 2005). "A novel function of transcription factor alpha-Pal/NRF-1: increasing neurite outgrowth". Biochem. Biophys. Res. Commun. 334 (1): 199–206. doi:10.1016/j.bbrc.2005.06.079. PMID 15992771.
- ^ "Entrez Gene: NRF1 nuclear respiratory factor 1". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4899.
- ^ Wang W, Kwok AM, Chan JY (August 2007). "The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription". J. Biol. Chem. 282 (34): 24670–8. doi:10.1074/jbc.M700159200. PMID 17609210.
- ^ Herzig RP, Andersson U, Scarpulla RC (December 2000). "Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and drosophila". J. Cell. Sci. 113 Pt 23: 4263–73. PMID 11069771.
- ^ Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM (July 1999). "Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1". Cell 98 (1): 115–24. doi:10.1016/S0092-8674(00)80611-X. PMID 10412986.
Further reading
- Tiranti V, Rossi E, Rocchi M et al. (1995). "The gene (NFE2L1) for human NRF-1, an activator involved in nuclear-mitochondrial interactions, maps to 7q32". Genomics 27 (3): 555–7. doi:10.1006/geno.1995.1094. PMID 7558044.
- Gopalakrishnan L, Scarpulla RC (1995). "Structure, expression, and chromosomal assignment of the human gene encoding nuclear respiratory factor 1". J. Biol. Chem. 270 (30): 18019–25. doi:10.1074/jbc.270.30.18019. PMID 7629110.
- Efiok BJ, Chiorini JA, Safer B (1994). "A key transcription factor for eukaryotic initiation factor-2 alpha is strongly homologous to developmental transcription factors and may link metabolic genes to cellular growth and development". J. Biol. Chem. 269 (29): 18921–30. PMID 8034649.
- Virbasius JV, Scarpulla RC (1994). "Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis". Proc. Natl. Acad. Sci. U.S.A. 91 (4): 1309–13. doi:10.1073/pnas.91.4.1309. PMC 43147. PMID 8108407. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=43147.
- Virbasius CA, Virbasius JV, Scarpulla RC (1994). "NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators". Genes Dev. 7 (12A): 2431–45. doi:10.1101/gad.7.12a.2431. PMID 8253388.
- Spelbrink JN, Van den Bogert C (1996). "The pre-mRNA of nuclear respiratory factor 1, a regulator of mitochondrial biogenesis, is alternatively spliced in human tissues and cell lines". Hum. Mol. Genet. 4 (9): 1591–6. doi:10.1093/hmg/4.9.1591. PMID 8541844.
- Johnsen O, Skammelsrud N, Luna L et al. (1996). "Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1". Nucleic Acids Res. 24 (21): 4289–97. doi:10.1093/nar/24.21.4289. PMC 146217. PMID 8932385. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=146217.
- Venugopal R, Jaiswal AK (1997). "Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene". Proc. Natl. Acad. Sci. U.S.A. 93 (25): 14960–5. doi:10.1073/pnas.93.25.14960. PMC 26245. PMID 8962164. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=26245.
- Chen S, Nagy PL, Zalkin H (1997). "Role of NRF-1 in bidirectional transcription of the human GPAT-AIRC purine biosynthesis locus". Nucleic Acids Res. 25 (9): 1809–16. doi:10.1093/nar/25.9.1809. PMC 146651. PMID 9108165. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=146651.
- Toki T, Itoh J, Kitazawa J et al. (1997). "Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif". Oncogene 14 (16): 1901–10. doi:10.1038/sj.onc.1201024. PMID 9150357.
- Marini MG, Chan K, Casula L et al. (1997). "hMAF, a small human transcription factor that heterodimerizes specifically with Nrf1 and Nrf2". J. Biol. Chem. 272 (26): 16490–7. doi:10.1074/jbc.272.26.16490. PMID 9195958.
- Gugneja S, Scarpulla RC (1997). "Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding". J. Biol. Chem. 272 (30): 18732–9. doi:10.1074/jbc.272.30.18732. PMID 9228045.
- Johnsen O, Murphy P, Prydz H, Kolsto AB (1998). "Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription". Nucleic Acids Res. 26 (2): 512–20. doi:10.1093/nar/26.2.512. PMC 147270. PMID 9421508. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=147270.
- Au HC, Scheffler IE (1998). "Promoter analysis of the human succinate dehydrogenase iron-protein gene--both nuclear respiratory factors NRF-1 and NRF-2 are required". Eur. J. Biochem. 251 (1–2): 164–74. doi:10.1046/j.1432-1327.1998.2510164.x. PMID 9492280.
- Miranda S, Foncea R, Guerrero J, Leighton F (1999). "Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells". Biochem. Biophys. Res. Commun. 258 (1): 44–9. doi:10.1006/bbrc.1999.0580. PMID 10222232.
- Li B, Holloszy JO, Semenkovich CF (1999). "Respiratory uncoupling induces delta-aminolevulinate synthase expression through a nuclear respiratory factor-1-dependent mechanism in HeLa cells". J. Biol. Chem. 274 (25): 17534–40. doi:10.1074/jbc.274.25.17534. PMID 10364186.
- Wu Z, Puigserver P, Andersson U et al. (1999). "Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1". Cell 98 (1): 115–24. doi:10.1016/S0092-8674(00)80611-X. PMID 10412986.
- Solecki D, Bernhardt G, Lipp M, Wimmer E (2000). "Identification of a nuclear respiratory factor-1 binding site within the core promoter of the human polio virus receptor/CD155 gene". J. Biol. Chem. 275 (17): 12453–62. doi:10.1074/jbc.275.17.12453. PMID 10777530.
- Kumari D, Usdin K (2001). "Interaction of the transcription factors USF1, USF2, and alpha -Pal/Nrf-1 with the FMR1 promoter. Implications for Fragile X mental retardation syndrome". J. Biol. Chem. 276 (6): 4357–64. doi:10.1074/jbc.M009629200. PMID 11058604.
External links
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) Miscellaneoussee also transcription factor/coregulator deficiencies
B bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°)Categories:- Human proteins
- Transcription factors
- Chromosome 7 gene stubs
Wikimedia Foundation. 2010.
