- NFATC2
-
Nuclear factor of activated T-cells, cytoplasmic 2 is a protein that in humans is encoded by the NFATC2 gene.[1]
This gene is a member of the nuclear factor of activated T cells (NFAT) family. The product of this gene is a DNA-binding protein with a REL-homology region (RHR) and an NFAT-homology region (NHR). This protein is present in the cytosol and only translocates to the nucleus upon T cell receptor (TCR) stimulation, where it becomes a member of the nuclear factors of activated T cells transcription complex. This complex plays a central role in inducing gene transcription during the immune response. Alternate transcriptional splice variants, encoding different isoforms, have been characterized.[2]
NFAT transcription factors are implicated in breast cancer, more specifically in the process of cell motility at the basis of metastasis formation. Indeed NFAT1 (NFATC2) is pro-invasive and pro-migratory in breast carcinoma [3] [4]Contents
Interactions
NFATC2 has been shown to interact with MEF2D,[5] EP300,[6] IRF4[7] and Protein kinase Mζ.[8]
See also
References
- ^ Northrop JP, Ho SN, Chen L, Thomas DJ, Timmerman LA, Nolan GP, Admon A, Crabtree GR (Jul 1994). "NF-AT components define a family of transcription factors targeted in T-cell activation". Nature 369 (6480): 497–502. doi:10.1038/369497a0. PMID 8202141.
- ^ "Entrez Gene: NFATC2 nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4773.
- ^ Jauliac, S; López-Rodriguez, C, Shaw, LM, Brown, LF, Rao, A, Toker, A (2002 Jul). "The role of NFAT transcription factors in integrin-mediated carcinoma invasion.". Nature cell biology 4 (7): 540–4. doi:10.1038/ncb816. PMID 12080349.
- ^ Yoeli-Lerner, M; Yiu, GK, Rabinovitz, I, Erhardt, P, Jauliac, S, Toker, A (2005 Nov 23). "Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT.". Molecular cell 20 (4): 539–50. PMID 16307918.
- ^ Youn, H D; Chatila T A, Liu J O (Aug. 2000). "Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis". EMBO J. (ENGLAND) 19 (16): 4323–4331. doi:10.1093/emboj/19.16.4323. ISSN 0261-4189. PMC 302027. PMID 10944115. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=302027.
- ^ García-Rodríguez, C; Rao A (Jun. 1998). "Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP)". J. Exp. Med. (UNITED STATES) 187 (12): 2031–2036. doi:10.1084/jem.187.12.2031. ISSN 0022-1007. PMC 2212364. PMID 9625762. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2212364.
- ^ Rengarajan, Jyothi; Mowen Kerri A, McBride Kathryn D, Smith Erica D, Singh Harinder, Glimcher Laurie H (Apr. 2002). "Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression". J. Exp. Med. (United States) 195 (8): 1003–1012. doi:10.1084/jem.20011128. ISSN 0022-1007. PMC 2193700. PMID 11956291. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2193700.
- ^ San-Antonio, Belén; Iñiguez Miguel A, Fresno Manuel (Jul. 2002). "Protein kinase Czeta phosphorylates nuclear factor of activated T cells and regulates its transactivating activity". J. Biol. Chem. (United States) 277 (30): 27073–27080. doi:10.1074/jbc.M106983200. ISSN 0021-9258. PMID 12021260.
Further reading
- Rao A, Luo C, Hogan PG (1997). "Transcription factors of the NFAT family: regulation and function". Annu. Rev. Immunol. 15: 707–747. doi:10.1146/annurev.immunol.15.1.707. PMID 9143705.
- Crabtree GR (1999). "Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT". Cell 96 (5): 611–614. doi:10.1016/S0092-8674(00)80571-1. PMID 10089876.
- Horsley V, Pavlath GK (2002). "NFAT: ubiquitous regulator of cell differentiation and adaptation". J. Cell Biol. 156 (5): 771–774. doi:10.1083/jcb.200111073. PMC 2173310. PMID 11877454. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2173310.
- Aramburu J, Azzoni L, Rao A, Perussia B (1995). "Activation and expression of the nuclear factors of activated T cells, NFATp and NFATc, in human natural killer cells: regulation upon CD16 ligand binding". J. Exp. Med. 182 (3): 801–810. doi:10.1084/jem.182.3.801. PMC 2192167. PMID 7650486. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2192167.
- Auffray C, Behar G, Bois F et al. (1995). "[IMAGE: molecular integration of the analysis of the human genome and its expression]". C. R. Acad. Sci. III, Sci. Vie 318 (2): 263–72. PMID 7757816.
- Li X, Ho SN, Luna J et al. (1995). "Cloning and chromosomal localization of the human and murine genes for the T-cell transcription factors NFATc and NFATp". Cytogenet. Cell Genet. 68 (3–4): 185–191. doi:10.1159/000133910. PMID 7842733.
- Ho S, Timmerman L, Northrop J, Crabtree GR (1995). "Cloning and characterization of NF-ATc and NF-ATp: the cytoplasmic components of NF-AT". Adv. Exp. Med. Biol. 365: 167–73. PMID 7887301.
- Jabado N, Le Deist F, Fisher A, Hivroz C (1994). "Interaction of HIV gp120 and anti-CD4 antibodies with the CD4 molecule on human CD4+ T cells inhibits the binding activity of NF-AT, NF-kappa B and AP-1, three nuclear factors regulating interleukin-2 gene enhancer activity". Eur. J. Immunol. 24 (11): 2646–2652. doi:10.1002/eji.1830241112. PMID 7957556.
- Vacca A, Farina M, Maroder M et al. (1995). "Human immunodeficiency virus type-1 tat enhances interleukin-2 promoter activity through synergism with phorbol ester and calcium-mediated activation of the NF-AT cis-regulatory motif". Biochem. Biophys. Res. Commun. 205 (1): 467–474. doi:10.1006/bbrc.1994.2689. PMID 7999066.
- Jain J, McCaffrey PG, Miner Z et al. (1993). "The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun". Nature 365 (6444): 352–355. doi:10.1038/365352a0. PMID 8397339.
- Luo C, Burgeon E, Carew JA et al. (1996). "Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes". Mol. Cell. Biol. 16 (7): 3955–66. PMC 231392. PMID 8668213. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=231392.
- Di Somma MM, Majolini MB, Burastero SE et al. (1996). "Cyclosporin A sensitivity of the HIV-1 long terminal repeat identifies distinct p56lck-dependent pathways activated by CD4 triggering". Eur. J. Immunol. 26 (9): 2181–2188. doi:10.1002/eji.1830260933. PMID 8814265.
- Copeland KF, McKay PJ, Rosenthal KL (1996). "Suppression of the human immunodeficiency virus long terminal repeat by CD8+ T cells is dependent on the NFAT-1 element". AIDS Res. Hum. Retroviruses 12 (2): 143–148. doi:10.1089/aid.1996.12.143. PMID 8834464.
- Bonaldo MF, Lennon G, Soares MB (1997). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Res. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Hodge MR, Chun HJ, Rengarajan J et al. (1997). "NF-AT-Driven interleukin-4 transcription potentiated by NIP45". Science 274 (5294): 1903–1905. doi:10.1126/science.274.5294.1903. PMID 8943202.
- Lyakh L, Ghosh P, Rice NR (1997). "Expression of NFAT-family proteins in normal human T cells". Mol. Cell. Biol. 17 (5): 2475–84. PMC 232096. PMID 9111316. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=232096.
- Amasaki Y, Masuda ES, Imamura R et al. (1998). "Distinct NFAT family proteins are involved in the nuclear NFAT-DNA binding complexes from human thymocyte subsets". J. Immunol. 160 (5): 2324–33. PMID 9498773.
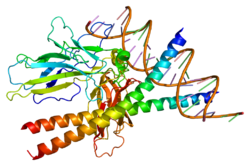
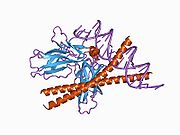
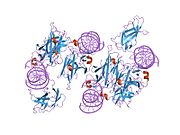
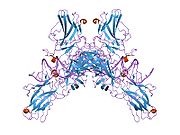
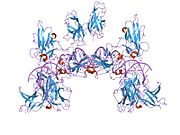
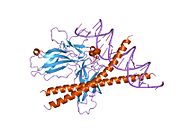
- Chen L, Glover JN, Hogan PG et al. (1998). "Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA". Nature 392 (6671): 42–48. doi:10.1038/32100. PMID 9510247.
PDB gallery 1a02: STRUCTURE OF THE DNA BINDING DOMAINS OF NFAT, FOS AND JUN BOUND TO DNA1owr: CRYSTAL STRUCTURE OF HUMAN NFAT1 BOUND MONOMERICALLY TO DNA1p7h: Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element1pzu: An asymmetric NFAT1-RHR homodimer on a pseudo-palindromic, Kappa-B site1s9k: Crystal Structure of Human NFAT1 and Fos-Jun on the IL-2 ARRE1 Site2as5: Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA.External links
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) MiscellaneousCategories:- Human proteins
- Transcription factors
- Chromosome 20 gene stubs
Wikimedia Foundation. 2010.