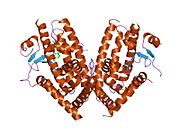
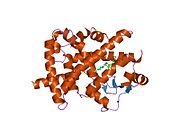
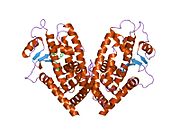
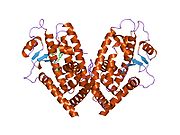
- Peroxisome proliferator-activated receptor gamma
-
Peroxisome proliferator-activated receptor gamma (PPAR-γ or PPARG), also known as the glitazone receptor, or NR1C3 (nuclear receptor subfamily 1, group C, member 3) is a type II nuclear receptor that in humans is encoded by the PPARG gene.[1][2][3]
Two isoforms of PPARG are detected in the human and in the mouse: PPAR-γ1 (found in nearly all tissues except muscle) and PPAR-γ2 (mostly found in adipose tissue and the intestine).[4]
Contents
Function
PPARG regulates fatty acid storage and glucose metabolism. The genes activated by PPARG stimulate lipid uptake and adipogenesis by fat cells. PPARG knockout mice fail to generate adipose tissue when fed a high fat diet.[5]
This gene encodes a member of the peroxisome proliferator-activated receptor (PPAR) subfamily of nuclear receptors. PPARs form heterodimers with retinoid X receptors (RXRs) and these heterodimers regulate transcription of various genes. Three subtypes of PPARs are known: PPAR-alpha, PPAR-delta, and PPAR-gamma. The protein encoded by this gene is PPAR-gamma and is a regulator of adipocyte differentiation. Alternatively spliced transcript variants that encode different isoforms have been described.[6]
Interactions
Peroxisome proliferator-activated receptor gamma has been shown to interact with:
Clinical relevance
PPAR-gamma has been implicated in the pathology of numerous diseases including obesity, diabetes, atherosclerosis and cancer. PPAR-gamma agonists have been used in the treatment of dyslipidaemia and hyperglycemia.[17] PPAR-gamma decreases the inflammatory response of many cardiovascular cells, particularly endothelial cells.[18] PPAR-gamma activates the PON1 gene, increasing synthesis and release of paraoxonase 1 from the liver, reducing atherosclerosis.[19]
Many insulin sensitizing drugs used in the treatment of diabetes target PPARG as a means to lower serum glucose without increasing pancreatic insulin secretion.
A fusion protein of PPAR-γ1 and the thyroid transcription factor PAX8 is present in approximately one-third of follicular thyroid carcinomas, specifically those cancers with a chromosomal translocation of t(2;3)(q13;p25), which permits juxtaposition of portions of both genes.[20] [21]References
- ^ Greene ME, Blumberg B, McBride OW, Yi HF, Kronquist K, Kwan K, Hsieh L, Greene G, Nimer SD (1995). "Isolation of the human peroxisome proliferator activated receptor gamma cDNA: expression in hematopoietic cells and chromosomal mapping". Gene Expr. 4 (4–5): 281–99. PMID 7787419.
- ^ Elbrecht A, Chen Y, Cullinan CA, Hayes N, Leibowitz M, Moller DE, Berger J (July 1996). "Molecular cloning, expression and characterization of human peroxisome proliferator activated receptors gamma 1 and gamma 2". Biochem. Biophys. Res. Commun. 224 (2): 431–7. doi:10.1006/bbrc.1996.1044. PMID 8702406.
- ^ Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, Palmer CN, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W (December 2006). "International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors". Pharmacol. Rev. 58 (4): 726–41. doi:10.1124/pr.58.4.5. PMID 17132851.
- ^ Fajas L, Auboeuf D, Raspé E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J (July 1997). "The organization, promoter analysis, and expression of the human PPARgamma gene". J. Biol. Chem. 272 (30): 18779–89. doi:10.1074/jbc.272.30.18779. PMID 9228052.
- ^ Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB, Magnuson MA (April 2005). "Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance". Proc. Natl. Acad. Sci. U.S.A. 102 (17): 6207–12. doi:10.1073/pnas.0306743102. PMC 556131. PMID 15833818. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=556131.
- ^ "Entrez Gene: PPARG peroxisome proliferator-activated receptor gamma". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5468.
- ^ Brendel C, Gelman L, Auwerx J (June 2002). "Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism". Mol. Endocrinol. 16 (6): 1367–77. doi:10.1210/me.16.6.1367. PMID 12040021.
- ^ Berger J, Patel HV, Woods J, Hayes NS, Parent SA, Clemas J, Leibowitz MD, Elbrecht A, Rachubinski RA, Capone JP, Moller DE (April 2000). "A PPARgamma mutant serves as a dominant negative inhibitor of PPAR signaling and is localized in the nucleus". Mol. Cell. Endocrinol. 162 (1–2): 57–67. doi:10.1016/S0303-7207(00)00211-2. PMID 10854698.
- ^ Gampe RT, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE (March 2000). "Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors". Mol. Cell 5 (3): 545–55. doi:10.1016/S1097-2765(00)80448-7. PMID 10882139.
- ^ a b c Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J (December 2002). "The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation". Dev. Cell 3 (6): 903–10. doi:10.1016/S1534-5807(02)00360-X. PMID 12479814.
- ^ a b c d Kodera Y, Takeyama K, Murayama A, Suzawa M, Masuhiro Y, Kato S (October 2000). "Ligand type-specific interactions of peroxisome proliferator-activated receptor gamma with transcriptional coactivators". J. Biol. Chem. 275 (43): 33201–4. doi:10.1074/jbc.C000517200. PMID 10944516.
- ^ Franco PJ, Li G, Wei LN (August 2003). "Interaction of nuclear receptor zinc finger DNA binding domains with histone deacetylase". Mol. Cell. Endocrinol. 206 (1–2): 1–12. doi:10.1016/S0303-7207(03)00254-5. PMID 12943985.
- ^ Heinlein CA, Ting HJ, Yeh S, Chang C (June 1999). "Identification of ARA70 as a ligand-enhanced coactivator for the peroxisome proliferator-activated receptor gamma". J. Biol. Chem. 274 (23): 16147–52. doi:10.1074/jbc.274.23.16147. PMID 10347167.
- ^ Nishizawa H, Yamagata K, Shimomura I, Takahashi M, Kuriyama H, Kishida K, Hotta K, Nagaretani H, Maeda N, Matsuda M, Kihara S, Nakamura T, Nishigori H, Tomura H, Moore DD, Takeda J, Funahashi T, Matsuzawa Y (January 2002). "Small heterodimer partner, an orphan nuclear receptor, augments peroxisome proliferator-activated receptor gamma transactivation". J. Biol. Chem. 277 (2): 1586–92. doi:10.1074/jbc.M104301200. PMID 11696534.
- ^ Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG (November 2003). "Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha". Mol. Cell 12 (5): 1137–49. doi:10.1016/S1097-2765(03)00391-5. PMID 14636573.
- ^ Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM (November 1999). "Activation of PPARgamma coactivator-1 through transcription factor docking". Science 286 (5443): 1368–71. doi:10.1126/science.286.5443.1368. PMID 10558993.
- ^ Li Y, Qi Y, Huang TH, Yamahara J, Roufogalis BD (January 2008). "Pomegranate flower: a unique traditional antidiabetic medicine with dual PPAR-alpha/-gamma activator properties". Diabetes Obes Metab 10 (1): 10–7. doi:10.1111/j.1463-1326.2007.00708.x. PMID 18095947.
- ^ Hamblin M, Chang L, Fan Y, Zhang J, Chen YE (June 2009). "PPARs and the Cardiovascular System". Antioxid. Redox Signal. 11 (6): 1415–52. doi:10.1089/ARS.2008.2280. PMC 2737093. PMID 19061437. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2737093.
- ^ Khateeb J, Gantman A, Kreitenberg AJ, Aviram M, Fuhrman B (January 2010). "Paraoxonase 1 (PON1) expression in hepatocytes is upregulated by pomegranate polyphenols: a role for PPAR-gamma pathway". Atherosclerosis 208 (1): 119–25. doi:10.1016/j.atherosclerosis.2009.08.051. PMID 19783251.
- ^ Kroll, TG; et al. (25). "PAX8-PPARγ1 Fusion in Oncogene Human Thyroid Carcinoma". Science 28 (5483): 1357–1360. doi:10.1126/science.289.5483.1357. http://www.sciencemag.org/content/289/5483/1357.abstract. Retrieved 8 December 2010.
- ^ Chapter 20 in: Mitchell, Richard Sheppard; Kumar, Vinay; Abbas, Abul K.; Fausto, Nelson. Robbins Basic Pathology. Philadelphia: Saunders. ISBN 1-4160-2973-7. 8th edition.
Further reading
- Qi C, Zhu Y, Reddy JK (2001). "Peroxisome proliferator-activated receptors, coactivators, and downstream targets". Cell Biochem. Biophys. 32 Spring: 187–204. PMID 11330046.
- Kadowaki T, Hara K, Kubota N, et al. (2002). "The role of PPARgamma in high-fat diet-induced obesity and insulin resistance". J. Diabetes Complicat. 16 (1): 41–5. doi:10.1016/S1056-8727(01)00206-9. PMID 11872365.
- Wakino S, Law RE, Hsueh WA (2002). "Vascular protective effects by activation of nuclear receptor PPARgamma". J. Diabetes Complicat. 16 (1): 46–9. doi:10.1016/S1056-8727(01)00197-0. PMID 11872366.
- Takano H, Komuro I (2002). "Roles of peroxisome proliferator-activated receptor gamma in cardiovascular disease". J. Diabetes Complicat. 16 (1): 108–14. doi:10.1016/S1056-8727(01)00203-3. PMID 11872377.
- Stumvoll M, Häring H (2002). "The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism". Diabetes 51 (8): 2341–7. doi:10.2337/diabetes.51.8.2341. PMID 12145143.
- Koeffler HP (2003). "Peroxisome proliferator-activated receptor gamma and cancers". Clin. Cancer Res. 9 (1): 1–9. PMID 12538445.
- Puigserver P, Spiegelman BM (2003). "Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator". Endocr. Rev. 24 (1): 78–90. doi:10.1210/er.2002-0012. PMID 12588810.
- Takano H, Hasegawa H, Nagai T, Komuro I (2003). "The role of PPARgamma-dependent pathway in the development of cardiac hypertrophy". Drugs Today 39 (5): 347–57. doi:10.1358/dot.2003.39.5.799458. PMID 12861348.
- Rangwala SM, Lazar MA (2004). "Peroxisome proliferator-activated receptor gamma in diabetes and metabolism". Trends Pharmacol. Sci. 25 (6): 331–6. doi:10.1016/j.tips.2004.03.012. PMID 15165749.
- Cuzzocrea S (2005). "Peroxisome proliferator-activated receptors gamma ligands and ischemia and reperfusion injury". Vascul. Pharmacol. 41 (6): 187–95. doi:10.1016/j.vph.2004.10.004. PMID 15653094.
- Savage DB (2007). "PPAR gamma as a metabolic regulator: insights from genomics and pharmacology". Expert reviews in molecular medicine 7 (1): 1–16. doi:10.1017/S1462399405008793. PMID 15673477.
- Pégorier JP (2005). "[PPAR receptors and insulin sensitivity: new agonists in development]". Ann. Endocrinol. (Paris) 66 (2 Pt 2): 1S10–7. PMID 15959400.
- Tsai YS, Maeda N (2005). "PPARgamma: a critical determinant of body fat distribution in humans and mice". Trends Cardiovasc. Med. 15 (3): 81–5. doi:10.1016/j.tcm.2005.04.002. PMID 16039966.
- Gurnell M (2006). "Peroxisome proliferator-activated receptor gamma and the regulation of adipocyte function: lessons from human genetic studies". Best Pract. Res. Clin. Endocrinol. Metab. 19 (4): 501–23. doi:10.1016/j.beem.2005.10.001. PMID 16311214.
- Cecil JE, Watt P, Palmer CN, Hetherington M (2006). "Energy balance and food intake: the role of PPARgamma gene polymorphisms". Physiol. Behav. 88 (3): 227–33. doi:10.1016/j.physbeh.2006.05.028. PMID 16777151.
- Rousseaux C, Desreumaux P (2007). "[The peroxisome-proliferator-activated gamma receptor and chronic inflammatory bowel disease (PPARgamma and IBD)]". J. Soc. Biol. 200 (2): 121–31. doi:10.1051/jbio:2006015. PMID 17151549.
- Eriksson JG (2007). "Gene polymorphisms, size at birth, and the development of hypertension and type 2 diabetes". J. Nutr. 137 (4): 1063–5. PMID 17374678.
- Tönjes A, Stumvoll M (2007). "The role of the Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma in diabetes risk". Current opinion in clinical nutrition and metabolic care 10 (4): 410–4. doi:10.1097/MCO.0b013e3281e389d9. PMID 17563457.
- Burgermeister E, Seger R (2007). "MAPK kinases as nucleo-cytoplasmic shuttles for PPARgamma". Cell Cycle 6 (13): 1539–48. doi:10.4161/cc.6.13.4453. PMID 17611413.
- Papageorgiou E, Pitulis N, Msaouel P, et al. (2007). "The non-genomic crosstalk between PPAR-gamma ligands and ERK1/2 in cancer cell lines". Expert Opin. Ther. Targets 11 (8): 1071–85. doi:10.1517/14728222.11.8.1071. PMID 17665979.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
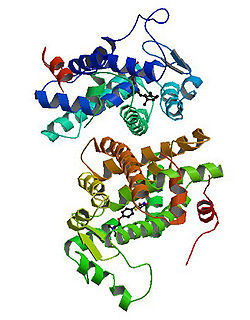
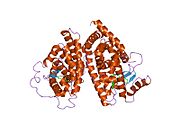
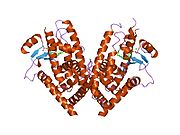
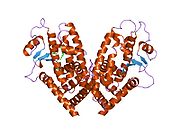
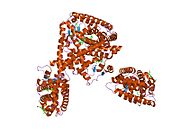
PDB gallery 1fm6: THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND ROSIGLITAZONE AND CO-ACTIVATOR PEPTIDES.1fm9: THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND GI262570 AND CO-ACTIVATOR PEPTIDES.1i7i: CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF HUMAN PPAR-GAMMA IN COMPLEX WITH THE AGONIST AZ 2421k74: The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides.1knu: LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA IN COMPLEX WITH A SYNTHETIC AGONIST1nyx: Ligand binding domain of the human peroxisome proliferator activated receptor gamma in complex with an agonist1prg: LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA1rdt: Crystal Structure of a new rexinoid bound to the RXRalpha ligand binding doamin in the RXRalpha/PPARgamma heterodimer1wm0: PPARgamma in complex with a 2-BABA compound1zeo: Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist1zgy: Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP2ath: Crystal structure of the ligand binding domain of human PPAR-gamma im complex with an agonist2f4b: Crystal structure of the ligand binding domain of human PPAR-gamma in complex with an agonist2fvj: A novel anti-adipogenic partial agonist of peroxisome proliferator-activated receptor-gamma (PPARG) recruits pparg-coactivator-1 alpha (PGC1A) but potentiates insulin signaling in vitro2g0g: Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities2g0h: Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities2gtk: Structure-based Design of Indole Propionic Acids as Novel PPARag CO-Agonists2hfp: Crystal Structure of PPAR Gamma with N-sulfonyl-2-indole carboxamide ligands2i4j: Crystal structure of the complex between PPARgamma and the agonist LT160 (ureidofibrate derivative)2i4p: Crystal structure of the complex between PPARgamma and the partial agonist LT127 (ureidofibrate derivative). Structure obtained from crystals of the apo-form soaked for 30 days.2i4z: Crystal structure of the complex between PPARgamma and the partial agonist LT127 (ureidofibrate derivative). This structure has been obtained from crystals soaked for 6 hours.2om9: Ajulemic acid, a synthetic cannabinoid bound to PPAR gamma2prg: LIGAND-BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA3prg: LIGAND BINDING DOMAIN OF HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR4prg: 0072 PARTIAL AGONIST PPAR GAMMA COCRYSTALTranscription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) MiscellaneousCategories:- Human proteins
- Intracellular receptors
- Transcription factors
Wikimedia Foundation. 2010.