- Notch signaling pathway
-
The notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms.[1] Notch is present in all metazoans, and mammals possess four different notch receptors, referred to as NOTCH1, NOTCH2, NOTCH3, and NOTCH4. The notch receptor is a single-pass transmembrane receptor protein. It is a hetero-oligomer composed of a large extracellular portion, which associates in a calcium-dependent, non-covalent interaction with a smaller piece of the notch protein composed of a short extracellular region, a single transmembrane-pass, and a small intracellular region.[2]. Notch signaling promotes proliferative signaling during neurogenesis and it's activity is inhibited by Numb to promote neural differentiation.
Contents
Discovery
In 1914, John S. Dexter noticed the appearance of a notch in the wings of the fruit fly Drosophila melanogaster. The alleles of the gene were identified in 1917 by Thomas Hunt Morgan.[3][4] Its molecular analysis and sequencing was independently undertaken in the 1980s by Spyros Artavanis-Tsakonas and Michael W. Young.[5][6]
Mechanism of action
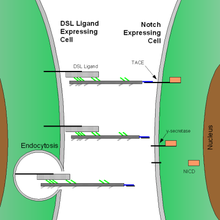
Further information: Notch proteinThe Notch protein sits like a trigger spanning the cell membrane, with part of it inside and part outside. Ligand proteins binding to the extracellular domain induce proteolytic cleavage and release of the intracellular domain, which enters the cell nucleus to modify gene expression.[7]
Because most ligands are also transmembrane proteins, the receptor is normally triggered only from direct cell-to-cell contact. In this way, groups of cells can organise themselves, such that, if one cell expresses a given trait, this may be switched off in neighbour cells by the intercellular notch signal. In this way, groups of cells influence one another to make large structures. Thus, lateral inhibition mechanisms are key to Notch signaling.
The notch cascade consists of notch and notch ligands, as well as intracellular proteins transmitting the notch signal to the cell's nucleus. The Notch/Lin-12/Glp-1 receptor family[8] was found to be involved in the specification of cell fates during development in Drosophila and C. elegans.[9]
Function
The notch signaling pathway is important for cell-cell communication, which involves gene regulation mechanisms that control multiple cell differentiation processes during embryonic and adult life. Notch signaling also has a role in the following processes:
-
- neuronal function and development[10][11][12][13]
- stabilization of arterial endothelial fate and angiogenesis[14]
- regulation of crucial cell communication events between endocardium and myocardium during both the formation of the valve primordial and ventricular development and differentiation[15]
- cardiac valve homeostasis, as well as implications in other human disorders involving the cardiovascular system[16]
- timely cell lineage specification of both endocrine and exocrine pancreas[17]
- influencing of binary fate decisions of cells that must choose between the secretory and absorptive lineages in the gut[18]
- expansion of the hematopoietic stem cell compartment during bone development and participation in commitment to the osteoblastic lineage, suggesting a potential therapeutic role for notch in bone regeneration and osteoporosis[19]
- T cell lineage commitment from common lymphoid precursor [20]
- regulation of cell-fate decision in mammary glands at several distinct development stages[21]
- possibly some non-nuclear mechanisms, such as control of the actin cytoskeleton through the tyrosine kinase Abl[22]
Notch signaling is dysregulated in many cancers,[23] and faulty notch signaling is implicated in many diseases including T-ALL (T-cell acute lymphoblastic leukemia),[24] CADASIL (Cerebral Autosomal Dominant Arteriopathy with Sub-cortical Infarcts and Leukoencephalopathy), MS (Multiple Sclerosis), Tetralogy of Fallot, Alagille syndrome, and many other disease states.
Inhibition of notch signaling has been shown to have anti-proliferative effects on T-ALL in cultured cells and in a mouse model.[25][26][27]
Pathway
Maturation of the notch receptor involves cleavage at the prospective extracellular side during intracellular trafficking in the Golgi complex.[28] This results in a bipartite protein, composed of a large extracellular domain linked to the smaller transmembrane and intracellular domain. Binding of ligand promotes two proteolytic processing events; as a result of proteolysis, the intracellular domain is liberated and can enter the nucleus to engage other DNA-binding proteins and regulate gene expression.
Notch and most of its ligands are transmembrane proteins, so the cells expressing the ligands typically must be adjacent to the notch expressing cell for signaling to occur.[citation needed] The notch ligands are also single-pass transmembrane proteins and are members of the DSL (Delta/Serrate/LAG-2) family of proteins. In Drosophila melanogaster (the fruit fly), there are two ligands named Delta and Serrate. In mammals, the corresponding names are Delta-like and Jagged. In mammals there are multiple Delta-like and Jagged ligands, as well as possibly a variety of other ligands, such as F3/contactin.[22]
In the nematode Caenorhabditis elegans, two genes encode homologous proteins, glp-1 and lin-12. There has been at least one report that suggests that some cells can send out processes that allow signaling to occur between cells that are as much as four or five cell diameters apart.[citation needed]
The notch extracellular domain is composed primarily of small cysteine knot motifs called EGF-like repeats.[29]
Notch 1, for example, has 36 of these repeats. Each EGF-like repeat is composed of approximately 40 amino acids, and its structure is defined largely by six conserved cysteine residues that form three conserved disulfide bonds. Each EGF-like repeat can be modified by O-linked glycans at specific sites.[30] An O-glucose sugar may be added between the first and second conserved cysteines, and an O-fucose may be added between the second and third conserved cysteines. These sugars are added by an as-yet-unidentified O-glucosyltransferase, and GDP-fucose Protein O-fucosyltransferase 1 (POFUT1), respectively. The addition of O-fucose by POFUT1 is absolutely necessary for notch function, and, without the enzyme to add O-fucose, all notch proteins fail to function properly. As yet, the manner by which the glycosylation of notch affects function is not completely understood.
The O-glucose on notch can be further elongated to a trisaccharide with the addition of two xylose sugars by xylosyltransferases, and the O-fucose can be elongated to a tetrasaccharide by the ordered addition of an N-acetylglucosamine (GlcNAc) sugar by an N-Acetylglucosaminyltransferase called Fringe, the addition of a galactose by a galactosyltransferase, and the addition of a sialic acid by a sialyltransferase.[31]
To add another level of complexity, in mammals there are three Fringe GlcNAc-transferases, named lunatic fringe, manic fringe, and radical fringe. These enzymes are responsible for something called a "fringe effect" on notch signaling.[32] If Fringe adds a GlcNAc to the O-fucose sugar, then the subsequent addition of a galactose and sialic acid will occur. In the presence of this tetrasaccharide, notch signals strongly when it interacts with the Delta ligand, but has markedly inhibited signaling when interacting with the Jagged ligand.[33] The means by which this addition of sugar inhibits signaling through one ligand, and potentiates signaling through another is not clearly understood.
Once the notch extracellular domain interacts with a ligand, an ADAM-family metalloprotease called TACE (Tumor Necrosis Factor Alpha Converting Enzyme) cleaves the notch protein just outside the membrane.[2] This releases the extracellular portion of notch, which continues to interact with the ligand. The ligand plus the notch extracellular domain is then endocytosed by the ligand-expressing cell. There may be signaling effects in the ligand-expressing cell after endocytosis; this part of notch signaling is a topic of active research. After this first cleavage, an enzyme called γ-secretase (which is implicated in Alzheimer's disease) cleaves the remaining part of the notch protein just inside the inner leaflet of the cell membrane of the notch-expressing cell. This releases the intracellular domain of the notch protein, which then moves to the nucleus, where it can regulate gene expression by activating the transcription factor CSL. It was originally thought that these CSL proteins suppressed Notch target transcription. However, further research showed that when the intracellular domain binds to the complex, it switches from a repressor to an activator of transcription.[34] Other proteins also participate in the intracellular portion of the notch signaling cascade.
External links
- Diagram: notch signaling pathway in Homo sapiens
- Diagram: Notch signaling in Drosophila
- Netpath - A curated resource of signal transduction pathways in humans
- MeSH Notch+Receptors
References
- ^ Artavanis-Tsakonas, Spyros et al. (1999-04-30). "Notch Signaling: Cell Fate Control and Signal Integration in Development (Review)". Science 284 (5415): 770–776. Bibcode 1999Sci...284..770A. doi:10.1126/science.284.5415.770. PMID 10221902.
- ^ a b Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A (February 2000). "A novel proteolytic cleavage involved in notch signaling: the role of the disintegrin-metalloprotease TACE". Mol. Cell 5 (2): 207–16. doi:10.1016/S1097-2765(00)80417-7. PMID 10882063.
- ^ Thomas Hunt Morgan (1917). "The theory of the gene". American Naturalist 51 (609): 513–544. doi:10.1086/279629
- ^ Morgan, Thomas (1928). The theory of the gene (revised ed.). Yale University Press. pp. 77–81. ISBN 0824013840. http://onlinebooks.library.upenn.edu/webbin/book/lookupid?key=olbp24097.
- ^ Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S (December 1985). "Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats". Cell 43 (3 Pt 2): 567–81. doi:10.1016/0092-8674(85)90229-6. PMID 3935325.
- ^ Kidd S, Kelley MR, Young MW (September 1986). "Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors". Mol. Cell. Biol. 6 (9): 3094–108. PMC 367044. PMID 3097517. http://mcb.asm.org/cgi/pmidlookup?view=long&pmid=3097517.
- ^ Oswald F, Täuber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM (November 2001). "p300 acts as a transcriptional coactivator for mammalian notch-1". Mol. Cell. Biol. 21 (22): 7761–74. doi:10.1128/MCB.21.22.7761-7774.2001. PMC 99946. PMID 11604511. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=99946.
- ^ Artavanis-Tsakonas S, Matsuno K, Fortini ME (April 1995). "Notch signaling". Science 268 (5208): 225–32. Bibcode 1995Sci...268..225A. doi:10.1126/science.7716513. PMID 7716513. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=7716513.
- ^ Singson A, Mercer KB, L'Hernault SW (April 1998). "The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization". Cell 93 (1): 71–9. doi:10.1016/S0092-8674(00)81147-2. PMID 9546393.
- ^ Gaiano N, Fishell G (2002). "The role of notch in promoting glial and neural stem cell fates". Annu. Rev. Neurosci. 25 (1): 471–90. doi:10.1146/annurev.neuro.25.030702.130823. PMID 12052917.
- ^ Bolós V, Grego-Bessa J, de la Pompa JL (May 2007). "Notch signaling in development and cancer". Endocr. Rev. 28 (3): 339–63. doi:10.1210/er.2006-0046. PMID 17409286.
- ^ Aguirre A, Rubio ME, Gallo V (2010). "Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal". Nature 467 (7313): 323–327. Bibcode 2010Natur.467..323A. doi:10.1038/nature09347. PMC 2941915. PMID 20844536. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2941915.
- ^ Hitoshi S et al. (April 2002). "Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells". Genes Dev. 16 (7): 846–858. doi:10.1101/gad.975202. PMC 186324. PMID 11937492. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=186324.
- ^ Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M (January 2003). "Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis". Mol. Cell. Biol. 23 (1): 14–25. doi:10.1128/MCB.23.1.14-25.2003. PMC 140667. PMID 12482957. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=140667.
- ^ Grego-Bessa J, Luna-Zurita L, del Monte G, Bolós V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Pérez-Pomares JM, de la Pompa JL (March 2007). "Notch signaling is essential for ventricular chamber development". Dev. Cell 12 (3): 415–29. doi:10.1016/j.devcel.2006.12.011. PMC 2746361. PMID 17336907. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2746361.
- ^ The notch signaling pathway in cardiac development and tissue homeostasis
- ^ Murtaugh LC, Stanger BZ, Kwan KM, Melton DA (December 2003). "Notch signaling controls multiple steps of pancreatic differentiation". Proc. Natl. Acad. Sci. U.S.A. 100 (25): 14920–5. Bibcode 2003PNAS..10014920M. doi:10.1073/pnas.2436557100. PMC 299853. PMID 14657333. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=299853.
- ^ Sander GR, Powell BC (April 2004). "Expression of notch receptors and ligands in the adult gut". J. Histochem. Cytochem. 52 (4): 509–16. PMID 15034002. http://www.jhc.org/cgi/content/full/52/4/509.
- ^ Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A (April 2005). "Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated notch1 signaling". J. Biol. Chem. 280 (16): 15842–8. doi:10.1074/jbc.M412891200. PMID 15695512.
- ^ Laky K, Fowlkes BJ (April 2008). "Notch signaling in CD4 and CD8 T cell development.". Current opinion in immunology 20 (2): 197–202. doi:10.1016/j.coi.2008.03.004. PMC 2475578. PMID 18434124. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2475578.
- ^ Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS (2004). "Role of notch signaling in cell-fate determination of human mammary stem/progenitor cells". Breast Cancer Res. 6 (6): R605–15. doi:10.1186/bcr920. PMC 1064073. PMID 15535842. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1064073.
- ^ a b Lai EC (March 2004). "Notch signaling: control of cell communication and cell fate". Development 131 (5): 965–73. doi:10.1242/dev.01074. PMID 14973298.
- ^ "The NOTCH pathway and Cancer". healthvalue.net. http://www.healthvalue.net/notch.html. Retrieved 2009-03-08.
- ^ Sharma VM, Draheim KM, Kelliher MA (April 2007). "The Notch1/c-Myc pathway in T cell leukemia". Cell Cycle 6 (8): 927–30. doi:10.4161/cc.6.8.4134. PMID 17404512. http://www.landesbioscience.com/journals/cc/abstract.php?id=4134.
- ^ Moellering RE, et al. (2009-11-12). "Direct inhibition of the NOTCH transcription factor complex". Nature 462 (7270): 182–188. Bibcode 2009Natur.462..182M. doi:10.1038/nature08543. PMC 2951323. PMID 19907488. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2951323.
- ^ Arora, Paramjit S.; Ansari, Aseem Z. (2009-11-12). "Chemical biology: A notch above other inhibitors". Nature 462 (7270): 171–173. Bibcode 2009Natur.462..171A. doi:10.1038/462171a. PMID 19907487.
- ^ Bagley, Katherine (2009-11-11). "New drug target for cancer". The Scientist. http://www.the-scientist.com/blog/display/56143/. Retrieved 2009-11-11.
- ^ Munro S, Freeman M (July 2000). "The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD". Curr. Biol. 10 (14): 813–20. doi:10.1016/S0960-9822(00)00578-9. PMID 10899003.
- ^ Ma B, Simala-Grant JL, Taylor DE (December 2006). "Fucosylation in prokaryotes and eukaryotes". Glycobiology 16 (12): 158R–184R. doi:10.1093/glycob/cwl040. PMID 16973733.
- ^ Shao L, Luo Y, Moloney DJ, Haltiwanger R (November 2002). "O-glycosylation of EGF repeats: identification and initial characterization of a UDP-glucose: protein O-glucosyltransferase". Glycobiology 12 (11): 763–70. doi:10.1093/glycob/cwf085. PMID 12460944.
- ^ Lu L, Stanley P (2006). "Roles of O-fucose glycans in notch signaling revealed by mutant mice". Meth. Enzymol. 417: 127–36. doi:10.1016/S0076-6879(06)17010-X. PMID 17132502.
- ^ Thomas GB, van Meyel DJ (February 2007). "The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression". Development 134 (3): 591–600. doi:10.1242/dev.02754. PMID 17215308.
- ^ LaVoie MJ, Selkoe DJ (September 2003). "The notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments". J. Biol. Chem. 278 (36): 34427–37. doi:10.1074/jbc.M302659200. PMID 12826675.
- ^ Desbordes, Sabrina; Hernan Lopez-Schier (2005). "Drosophila Patterning: Delta-Notch Interactions". Encyclopedia of Life Sciences: 4. doi:10.1038/npg.els.0004194.
Signaling pathways Agents Intracellular signaling P+PsSignal transducing adaptor protein: Scaffold protein
2nd messenger: cAMP-dependent pathway · Ca2+ signaling · Lipid signaling · IP3/DAG pathwayBy location Other concepts Signaling pathway: notch signaling pathway Receptor on signaling cell Ligand Receptor on receiving cell Categories: -
Wikimedia Foundation. 2010.