- Mdm2
-
Mdm2 is an important negative regulator of the p53 tumor suppressor. It is the name of a gene as well as the protein encoded by that gene. Mdm2 protein functions both as an E3 ubiquitin ligase that recognizes the N-terminal trans-activation domain (TAD) of the p53 tumor suppressor and an inhibitor of p53 transcriptional activation.
Contents
Discovery and expression in tumor cells
The murine double minute (mdm2) oncogene, which codes for the Mdm2 protein, was originally cloned, along with two other genes (mdm1 and mdm3) from the transformed mouse cell line 3T3-DM. Mdm2 overexpression, in cooperation with oncogenic Ras, promotes transformation of primary rodent fibroblasts, and mdm2 expression led to tumor formation in nude mice. The human homologue of this protein was later identified and is sometimes called Hdm2. Further supporting the role of mdm2 as an oncogene, several human tumor types have been shown to have increased levels of Mdm2, including soft tissue sarcomas and osteosarcomas as well as breast tumors. An additional Mdm2 family member, Mdm4 (also called MdmX), has been discovered and is also an important negative regulator of p53.
Ubiquitination target: p53
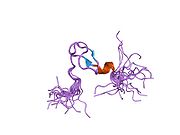
The key target of Mdm2 is the p53 tumor suppressor. Mdm2 has been identified as a p53 interacting protein that represses p53 transcriptional activity. Mdm2 achieves this repression by binding to and blocking the N-terminal trans-activation domain of p53. Mdm2 is a p53 responsive gene—that is, its transcription can be activated by p53. Thus when p53 is stabilized, the transcription of Mdm2 is also induced, resulting in higher Mdm2 protein levels.
E3 ligase activity
Mdm2 also acts as an E3 ubiquitin ligase, targeting both itself and p53 for degradation by the proteasome (see also Ubiquitin). Several lysine residues in p53 C-terminus have been identified as the sites of ubiquitination, and it has been shown that p53 protein levels are downregulated by Mdm2 in a proteasome-dependent manner. Mdm2 is capable of auto-polyubiquitination, and in complex with p300, a cooperating E3 ubiquitin ligase, is capable of polyubiquitinating p53. In this manner, Mdm2 and p53 are the members of a negative feedback control loop that keeps the level of p53 low in the absence of p53-stabilizing signals. This loop can be interfered with by kinases and genes like p14arf when p53 activation signals, including DNA damage, are high.
Structure and function
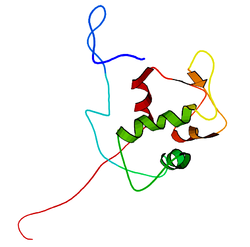
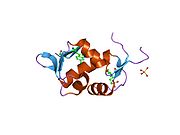
The full-length transcript of the mdm2 gene encodes a protein of 491 amino acids with a predicted molecular weight of 56kDa. This protein contains several conserved structural domains including an N-terminal p53 interaction domain, the structure of which has been solved using x-ray crystallography. The Mdm2 protein also contains a central acidic domain (residues 230-300). The phosphorylation of residues within this domain appears to be important for regulation of Mdm2 function. In addition, this region contains nuclear export and import signals that are essential for proper nuclear-cytoplasmic trafficking of Mdm2. Another conserved domain within the Mdm2 protein is a Zinc finger domain, the function of which is poorly understood.
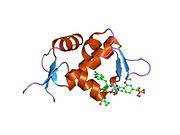
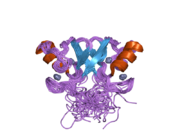
Mdm2 also contains a C-terminal RING domain (amino acid resdiues 430-480), which contains a Cis3-His2-Cis3 consensus that coordinates two molecules of zinc. These residues are required for zinc binding, which is essential for proper folding of the RING domain. The RING domain of Mdm2 confers E3 ubiquitin ligase activity and is sufficient for E3 ligase activity in Mdm2 RING autoubiquitination. The RING domain of Mdm2 is unique in that it incorporates a conserved Walker A or P-loop motif characteristic of nucleotide binding proteins, as well as a nucleolar localization sequence. The RING domain also binds specifically to RNA, although the function of this is poorly understood.
Regulation
There are several known mechanisms for regulation of Mdm2. One of these mechanisms is phosphorylation of the Mdm2 protein. Mdm2 is phosphorylated at multiple sites in cells. Following DNA damage, phosphorylation of Mdm2 leads to changes in protein function and stabilization of p53. Additionally, phosphorylation at certain residues within the central acidic domain of Mdm2 may stimulate its ability to target p53 for degradation. The induction of the p14arf protein, the alternate reading frame product of the p16INK4a locus, is also a mechanism of negatively regulating the p53-Mdm2 interaction. p14arf directly interacts with Mdm2 and leads to up-regulation of p53 transcriptional response. ARF sequesters Mdm2 in the nucleolus, resulting in inhibition of nuclear export and activation of p53, since nuclear export is essential for proper p53 degradation.
Inhibitors of the MDM2-p53 interaction include the cis-imidazoline analog nutlin.[2]
Levels and stability of Mdm2 are also modulated by ubiquitylation. Mdm2 auto ubiquitylates itself, which allows for its degradation by the proteasome. Mdm2 also interacts with a ubiquitin specific protease, USP7, which can reverse Mdm2-ubiquitylation and prevent it from being degraded by the proteasome. It is interesting to note that USP7 also protects from degradation the p53 protein, which is a major target of Mdm2. Thus Mdm2 and USP7 form an intricate circuit to finely regulate the stability and activity of p53, whose levels are critical for its function.
Interactions
Mdm2 has been shown to interact with HDAC1,[3] RPL26,[4] FKBP3,[5] CCNG1,[6] HTATIP,[7] GNL3,[8] Death associated protein 6,[9] PSME3,[10] Insulin-like growth factor 1 receptor,[11] RRM2B,[12] FOXO4,[13] Ribosomal protein L5,[14][15][8] Abl gene,[16] RYBP,[17] HIF1A,[18][19] PCAF,[20] TATA binding protein,[21][22] P73,[23][24] CTBP2,[25] NUMB,[26][27] P53,[28][29] P16,[14][30][31][32][9] PSMD10,[33] EP300,[34] CTBP1,[25] MDM4,[35][36][37][38] RPL11,[14][8] Promyelocytic leukemia protein,[39][40][41][42] Dihydrofolate reductase,[43] Arrestin beta 2,[44][45][46] Arrestin beta 1[45][46] and Ubiquitin C.[9][47][48]
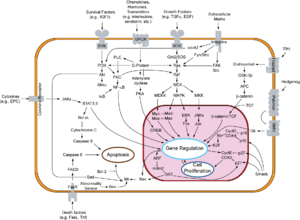 Overview of signal transduction pathways involved in apoptosis.
Overview of signal transduction pathways involved in apoptosis.
References
- ^ Uhrinova S, Uhrin D, Powers H, et al (2005). "Structure of free MDM2 N-terminal domain reveals conformational adjustments that accompany p53-binding". J. Mol. Biol. 350 (3): 587–98. doi:10.1016/j.jmb.2005.05.010. PMID 15953616.
- ^ Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004). "In vivo activation of the p53 pathway by small-molecule antagonists of MDM2". Science 6 (5659): 844–848. doi:10.1126/science.1092472. PMID 14704432.
- ^ Ito, Akihiro; Kawaguchi Yoshiharu, Lai Chun-Hsiang, Kovacs Jeffrey J, Higashimoto Yuichiro, Appella Ettore, Yao Tso-Pang (November 2002). "MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation". EMBO J. 21 (22): 6236–45. doi:10.1093/emboj/cdf616. PMC 137207. PMID 12426395. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=137207.
- ^ Ofir-Rosenfeld, Yaara; Boggs Kristy, Michael Dan, Kastan Michael B, Oren Moshe (October 2008). "Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26". Mol. Cell 32 (2): 180–9. doi:10.1016/j.molcel.2008.08.031. PMC 2587494. PMID 18951086. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2587494.
- ^ Ochocka, Anna Maria; Kampanis Petros, Nicol Samantha, Allende-Vega Nerea, Cox Miranda, Marcar Lynnette, Milne Diane, Fuller-Pace Frances, Meek David (February 2009). "FKBP25, a novel regulator of the p53 pathway, induces the degradation of MDM2 and activation of p53". FEBS Lett. 583 (4): 621–6. doi:10.1016/j.febslet.2009.01.009. PMID 19166840.
- ^ Zhao, Lili; Samuels Tina, Winckler Sarah, Korgaonkar Chandrashekhar, Tompkins Van, Horne Mary C, Quelle Dawn E (January 2003). "Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways". Mol. Cancer Res. 1 (3): 195–206. PMID 12556559.
- ^ Legube, Gaëlle; Linares Laetitia K, Lemercier Claudie, Scheffner Martin, Khochbin Saadi, Trouche Didier (April 2002). "Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation". EMBO J. (England) 21 (7): 1704–12. doi:10.1093/emboj/21.7.1704. PMC 125958. PMID 11927554. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=125958.
- ^ a b c Dai, Mu-Shui; Sun Xiao-Xin, Lu Hua (July 2008). "Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2". Mol. Cell. Biol. 28 (13): 4365–76. doi:10.1128/MCB.01662-07. PMC 2447154. PMID 18426907. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2447154.
- ^ a b c Ivanchuk, Stacey M; Mondal Soma, Rutka James T (June 2008). "p14ARF interacts with DAXX: effects on HDM2 and p53". Cell Cycle 7 (12): 1836–50. doi:10.4161/cc.7.12.6025. PMID 18583933.
- ^ Zhang, Zhuo; Zhang Ruiwen (March 2008). "Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation". EMBO J. 27 (6): 852–64. doi:10.1038/emboj.2008.25. PMC 2265109. PMID 18309296. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2265109.
- ^ Sehat, Bita; Andersson Sandra, Girnita Leonard, Larsson Olle (July 2008). "Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis". Cancer Res. 68 (14): 5669–77. doi:10.1158/0008-5472.CAN-07-6364. PMID 18632619.
- ^ Chang, Lufen; Zhou Bingsen, Hu Shuya, Guo Robin, Liu Xiyong, Jones Stephen N, Yen Yun (November 2008). "ATM-mediated serine 72 phosphorylation stabilizes ribonucleotide reductase small subunit p53R2 protein against MDM2 to DNA damage". Proc. Natl. Acad. Sci. U.S.A. 105 (47): 18519–24. doi:10.1073/pnas.0803313105. PMC 2587585. PMID 19015526. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2587585.
- ^ Brenkman, Arjan B; de Keizer Peter L J, van den Broek Niels J F, Jochemsen A G, Burgering Boudewijn M Th (2008). Cookson, Mark R.. ed. "Mdm2 induces mono-ubiquitination of FOXO4". PLoS ONE 3 (7): e2819. doi:10.1371/journal.pone.0002819. PMC 2475507. PMID 18665269. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2475507.
- ^ a b c Zhang, Yanping; Wolf Gabrielle White, Bhat Krishna, Jin Aiwen, Allio Theresa, Burkhart William A, Xiong Yue (December 2003). "Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway". Mol. Cell. Biol. 23 (23): 8902–12. doi:10.1128/MCB.23.23.8902-8912.2003. PMC 262682. PMID 14612427. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=262682.
- ^ Marechal, V; Elenbaas B, Piette J, Nicolas J C, Levine A J (November 1994). "The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes". Mol. Cell. Biol. 14 (11): 7414–20. PMC 359276. PMID 7935455. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=359276.
- ^ Goldberg, Zehavit; Vogt Sionov Ronit, Berger Michael, Zwang Yaara, Perets Ruth, Van Etten Richard A, Oren Moshe, Taya Yoichi, Haupt Ygal (July 2002). "Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation". EMBO J. 21 (14): 3715–27. doi:10.1093/emboj/cdf384. PMC 125401. PMID 12110584. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=125401.
- ^ Chen, Deng; Zhang Jianbing, Li Mao, Rayburn Elizabeth R, Wang Hui, Zhang Ruiwen (February 2009). "RYBP stabilizes p53 by modulating MDM2". EMBO Rep. 10 (2): 166–72. doi:10.1038/embor.2008.231. PMC 2637313. PMID 19098711. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2637313.
- ^ Chen, Delin; Li Muyang, Luo Jianyuan, Gu Wei (April 2003). "Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function". J. Biol. Chem. 278 (16): 13595–8. doi:10.1074/jbc.C200694200. PMID 12606552.
- ^ Ravi, R; Mookerjee B, Bhujwalla Z M, Sutter C H, Artemov D, Zeng Q, Dillehay L E, Madan A, Semenza G L, Bedi A (January 2000). "Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha". Genes Dev. 14 (1): 34–44. PMC 316350. PMID 10640274. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=316350.
- ^ Jin, Yetao; Zeng Shelya X, Dai Mu-Shui, Yang Xiang-Jiao, Lu Hua (August 2002). "MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation". J. Biol. Chem. 277 (34): 30838–43. doi:10.1074/jbc.M204078200. PMID 12068014.
- ^ Léveillard, T; Wasylyk B (December 1997). "The MDM2 C-terminal region binds to TAFII250 and is required for MDM2 regulation of the cyclin A promoter". J. Biol. Chem. 272 (49): 30651–61. doi:10.1074/jbc.272.49.30651. PMID 9388200.
- ^ Thut, C J; Goodrich J A, Tjian R (August 1997). "Repression of p53-mediated transcription by MDM2: a dual mechanism". Genes Dev. 11 (15): 1974–86. doi:10.1101/gad.11.15.1974. PMC 316412. PMID 9271120. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=316412.
- ^ Bálint, E; Bates S, Vousden K H (July 1999). "Mdm2 binds p73 alpha without targeting degradation". Oncogene 18 (27): 3923–9. doi:10.1038/sj.onc.1202781. PMID 10435614.
- ^ Zeng, X; Chen L, Jost C A, Maya R, Keller D, Wang X, Kaelin W G, Oren M, Chen J, Lu H (May 1999). "MDM2 suppresses p73 function without promoting p73 degradation". Mol. Cell. Biol. 19 (5): 3257–66. PMC 84120. PMID 10207051. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=84120.
- ^ a b Mirnezami, Alexander H; Campbell Sandra J, Darley Matthew, Primrose John N, Johnson Peter W M, Blaydes Jeremy P (July 2003). "Hdm2 recruits a hypoxia-sensitive corepressor to negatively regulate p53-dependent transcription". Curr. Biol. 13 (14): 1234–9. doi:10.1016/S0960-9822(03)00454-8. PMID 12867035.
- ^ Yogosawa, Satomi; Miyauchi Yasuhiro, Honda Reiko, Tanaka Hirofumi, Yasuda Hideyo (March 2003). "Mammalian Numb is a target protein of Mdm2, ubiquitin ligase". Biochem. Biophys. Res. Commun. 302 (4): 869–72. doi:10.1016/S0006-291X(03)00282-1. PMID 12646252.
- ^ Colaluca, Ivan N; Tosoni Daniela, Nuciforo Paolo, Senic-Matuglia Francesca, Galimberti Viviana, Viale Giuseppe, Pece Salvatore, Di Fiore Pier Paolo (January 2008). "NUMB controls p53 tumour suppressor activity". Nature 451 (7174): 76–80. doi:10.1038/nature06412. PMID 18172499.
- ^ Haupt Y, Maya R, Kazaz A, Oren M (May 1997). "Mdm2 promotes the rapid degradation of p53". Nature 387 (6630): 296–9. doi:10.1038/387296a0. PMID 9153395.
- ^ Honda, Tanaka & Yasuda 1997
- ^ Zhang Y, Xiong Y, Yarbrough WG (March 1998). "ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways". Cell 92 (6): 725–34. doi:10.1016/S0092-8674(00)81401-4. PMID 9529249.
- ^ Clark, Paula A; Llanos Susana, Peters Gordon (July 2002). "Multiple interacting domains contribute to p14ARF mediated inhibition of MDM2". Oncogene 21 (29): 4498–507. doi:10.1038/sj.onc.1205558. PMID 12085228.
- ^ Pomerantz, J; Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A (March 1998). "The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53". Cell 92 (6): 713–23. doi:10.1016/S0092-8674(00)81400-2. PMID 9529248.
- ^ Qiu W, Wu J, Walsh EM, et al. (July 2008). "Retinoblastoma protein modulates gankyrin-MDM2 in regulation of p53 stability and chemosensitivity in cancer cells". Oncogene 27 (29): 4034–43. doi:10.1038/onc.2008.43. PMID 18332869.
- ^ Grossman SR, Perez M, Kung AL, et al. (October 1998). "p300/MDM2 complexes participate in MDM2-mediated p53 degradation". Mol. Cell 2 (4): 405–15. doi:10.1016/S1097-2765(00)80140-9. PMID 9809062.
- ^ Kadakia, Madhavi; Brown Thomas L, McGorry Molly M, Berberich Steven J (December 2002). "MdmX inhibits Smad transactivation". Oncogene 21 (57): 8776–85. doi:10.1038/sj.onc.1205993. PMID 12483531.
- ^ Tanimura, S; Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M (March 1999). "MDM2 interacts with MDMX through their RING finger domains". FEBS Lett. 447 (1): 5–9. doi:10.1016/S0014-5793(99)00254-9. PMID 10218570.
- ^ Badciong, James C; Haas Arthur L (December 2002). "MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination". J. Biol. Chem. 277 (51): 49668–75. doi:10.1074/jbc.M208593200. PMID 12393902.
- ^ Linke, K; Mace P D, Smith C A, Vaux D L, Silke J, Day C L (May 2008). "Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans". Cell Death Differ. 15 (5): 841–8. doi:10.1038/sj.cdd.4402309. PMID 18219319.
- ^ Bernardi, Rosa; Scaglioni Pier Paolo, Bergmann Stephan, Horn Henning F, Vousden Karen H, Pandolfi Pier Paolo (July 2004). "PML regulates p53 stability by sequestering Mdm2 to the nucleolus". Nat. Cell Biol. 6 (7): 665–72. doi:10.1038/ncb1147. PMID 15195100.
- ^ Zhu, Hongyan; Wu Liqing, Maki Carl G (December 2003). "MDM2 and promyelocytic leukemia antagonize each other through their direct interaction with p53". J. Biol. Chem. 278 (49): 49286–92. doi:10.1074/jbc.M308302200. PMID 14507915.
- ^ Kurki S, Latonen L, Laiho M (October 2003). "Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization". J. Cell. Sci. 116 (Pt 19): 3917–25. doi:10.1242/jcs.00714. PMID 12915590.
- ^ Wei, Xiaolong; Yu Zhong Kang, Ramalingam Arivudainambi, Grossman Steven R, Yu Jiang H, Bloch Donald B, Maki Carl G (August 2003). "Physical and functional interactions between PML and MDM2". J. Biol. Chem. 278 (31): 29288–97. doi:10.1074/jbc.M212215200. PMID 12759344.
- ^ Maguire, Maria; Nield Paul C, Devling Timothy, Jenkins Rosalind E, Park B Kevin, Polański Radoslaw, Vlatković Nikolina, Boyd Mark T (May 2008). "MDM2 regulates dihydrofolate reductase activity through monoubiquitination". Cancer Res. 68 (9): 3232–42. doi:10.1158/0008-5472.CAN-07-5271. PMID 18451149.
- ^ Wang, Ping; Gao Hua, Ni Yanxiang, Wang Beibei, Wu Yalan, Ji Lili, Qin Linhua, Ma Lan, Pei Gang (February 2003). "Beta-arrestin 2 functions as a G-protein-coupled receptor-activated regulator of oncoprotein Mdm2". J. Biol. Chem. 278 (8): 6363–70. doi:10.1074/jbc.M210350200. PMID 12488444.
- ^ a b Wang, Ping; Wu Yalan, Ge Xin, Ma Lan, Pei Gang (March 2003). "Subcellular localization of beta-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus". J. Biol. Chem. 278 (13): 11648–53. doi:10.1074/jbc.M208109200. PMID 12538596.
- ^ a b Shenoy, Sudha K; Xiao Kunhong, Venkataramanan Vidya, Snyder Peter M, Freedman Neil J, Weissman Allan M (August 2008). "Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor". J. Biol. Chem. 283 (32): 22166–76. doi:10.1074/jbc.M709668200. PMC 2494938. PMID 18544533. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2494938.
- ^ Song, Min Sup; Song Su Jung, Kim So Yeon, Oh Hyun Jung, Lim Dae-Sik (July 2008). "The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex". EMBO J. 27 (13): 1863–74. doi:10.1038/emboj.2008.115. PMC 2486425. PMID 18566590. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2486425.
- ^ Yang, Wensheng; Dicker David T, Chen Jiandong, El-Deiry Wafik S (March 2008). "CARPs enhance p53 turnover by degrading 14-3-3sigma and stabilizing MDM2". Cell Cycle 7 (5): 670–82. doi:10.4161/cc.7.5.5701. PMID 18382127.
Further reading
- Cahilly-Snyder L, Yang-Feng T, Francke U, George DL (May 1987). "Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line". Somat. Cell Mol. Genet. 13 (3): 235–44. doi:10.1007/BF01535205. PMID 3474784.
- Chen J, Lin J, Levine AJ (January 1995). "Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene". Mol. Med. 1 (2): 142–52. PMC 2229942. PMID 8529093. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2229942.
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (March 2000). "Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53". J. Biol. Chem. 275 (12): 8945–51. doi:10.1074/jbc.275.12.8945. PMID 10722742. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=10722742.
- Freedman DA, Wu L, Levine AJ (January 1999). "Functions of the MDM2 oncoprotein". Cell. Mol. Life Sci. 55 (1): 96–107. doi:10.1007/s000180050273. PMID 10065155. http://link.springer.de/link/service/journals/00018/bibs/9055001/90550096.htm.
- Hay TJ, Meek DW (July 2000). "Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains". FEBS Lett. 478 (1-2): 183–6. doi:10.1016/S0014-5793(00)01850-0. PMID 10922493. http://linkinghub.elsevier.com/retrieve/pii/S0014-5793(00)01850-0.
- Honda, R; Tanaka, H; Yasuda, H (December 1997). "Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53". FEBS Lett. 420 (1): 25–7. doi:10.1016/S0014-5793(97)01480-4. PMID 9450543. http://linkinghub.elsevier.com/retrieve/pii/S0014-5793(97)01480-4.
- Honda R, Yasuda H (March 2000). "Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase". Oncogene 19 (11): 1473–6. doi:10.1038/sj.onc.1203464. PMID 10723139.
- Kubbutat MH, Jones SN, Vousden KH (May 1997). "Regulation of p53 stability by Mdm2". Nature 387 (6630): 299–303. doi:10.1038/387299a0. PMID 9153396.
- Kussie PH, Gorina S, Marechal V, et al. (November 1996). "Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain". Science 274 (5289): 948–53. doi:10.1126/science.274.5289.948. PMID 8875929. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=8875929.
- Meek DW, Knippschild U (December 2003). "Posttranslational modification of MDM2". Mol. Cancer Res. 1 (14): 1017–26. PMID 14707285. http://mcr.aacrjournals.org/cgi/pmidlookup?view=long&pmid=14707285.
- Midgley CA, Desterro JM, Saville MK, et al. (May 2000). "An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo". Oncogene 19 (19): 2312–23. doi:10.1038/sj.onc.1203593. PMID 10822382.
- Momand J, Wu HH, Dasgupta G (January 2000). "MDM2--master regulator of the p53 tumor suppressor protein". Gene 242 (1-2): 15–29. doi:10.1016/S0378-1119(99)00487-4. PMID 10721693. http://linkinghub.elsevier.com/retrieve/pii/S0378-1119(99)00487-4.
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ (June 1992). "The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation". Cell 69 (7): 1237–45. doi:10.1016/0092-8674(92)90644-R. PMID 1535557. http://linkinghub.elsevier.com/retrieve/pii/0092-8674(92)90644-R.
- Shieh SY, Ikeda M, Taya Y, Prives C (October 1997). "DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2". Cell 91 (3): 325–34. doi:10.1016/S0092-8674(00)80416-X. PMID 9363941. http://linkinghub.elsevier.com/retrieve/pii/S0092-8674(00)80416-X.
- Tao W, Levine AJ (June 1999). "P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2". Proc. Natl. Acad. Sci. U.S.A. 96 (12): 6937–41. doi:10.1073/pnas.96.12.6937. PMC 22020. PMID 10359817. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=10359817.
- Tao W, Levine AJ (March 1999). "Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53". Proc. Natl. Acad. Sci. U.S.A. 96 (6): 3077–80. doi:10.1073/pnas.96.6.3077. PMC 15897. PMID 10077639. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=10077639.
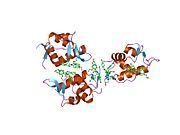
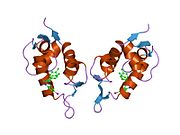
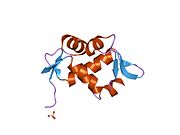
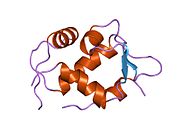
PDB gallery 1rv1: CRYSTAL STRUCTURE OF HUMAN MDM2 WITH AN IMIDAZOLINE INHIBITOR1t4e: Structure of Human MDM2 in complex with a Benzodiazepine Inhibitor1t4f: Structure of human MDM2 in complex with an optimized p53 peptide1ycr: MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P531z1m: NMR structure of unliganded MDM22axi: HDM2 in complex with a beta-hairpin2c6a: SOLUTION STRUCTURE OF THE C4 ZINC-FINGER DOMAIN OF HDM22c6b: SOLUTION STRUCTURE OF THE C4 ZINC-FINGER DOMAIN OF HDM22gv2: MDM2 in complex with an 8-mer p53 peptide analogue2hdp: Solution Structure of Hdm2 RING Finger DomainExternal links
Neoplasm: Tumor suppressor genes/proteins and Oncogenes/Proto-oncogenes Ligand Receptor TSP: CDH1TSP: PTCH1TSP: TGF beta receptor 2Intracellular signaling P+Ps ONCO: Beta-catenin · TSP: APCHippo signaling pathwayOther/unknownNucleus TSP: VHL · ONCO: CBL - MDM2Mitochondria Other/ungrouped M: NEO
tsoc, mrkr
tumr, epon, para
drug (L1i/1e/V03)
Enzymes: CO CS and CN ligases (EC 6.1-6.3) 6.1: Carbon-Oxygen Aminoacyl tRNA synthetase (Tyrosine, Tryptophan, Threonine, Leucine, Isoleucine, Lysine, Alanine, Valine, Methionine, Serine, Aspartate, D-alanine-poly(phosphoribitol) ligase, Glycine, Proline, Cysteine, Glutamate, Glutamine, Arginine, Phenylalanine, Histidine, Asparagine, Aspartate, Glutamate, Lysine)6.2: Carbon-Sulfur Succinyl coenzyme A synthetase - Acetyl Co-A synthetase - Long fatty acyl CoA synthetase6.3: Carbon-Nitrogen Glutamine synthetase - Ubiquitin ligase (Cullin, Von Hippel-Lindau tumor suppressor, UBE3A, Mdm2, Anaphase-promoting complex, UBR1) - Glutathione synthetase - CTP synthase - Adenylosuccinate synthase - Argininosuccinate synthetase - Holocarboxylase synthetase - GMP synthase - Asparagine synthetase - Carbamoyl phosphate synthetase (I, II) - Gamma-glutamylcysteine synthetaseLigases: carbon-carbon ligases (EC 6.4) Biotin dependent carboxylase Other 6.5: Phosphoric Ester 6.6: Nitrogen-Metal Chaperones/
protein foldingOtherProtein targeting Ubiquitin E1 Ubiquitin-activating enzyme (UBA1, UBA2, UBA3, UBA5, UBA6, UBA7, ATG7, NAE1, SAE1)
E2 Ubiquitin-conjugating enzyme (A • B • C • D1, D2, D3 • E1, E2, E3 • G1, G2 • H • I • J1, J2 • K • L1, L2, L3, L4, L6 • M • N • O • Q1, Q2 • R1 (CDC34), R2 • S • V1, V2 • Z)
E3 Ubiquitin ligase (VHL, Cullin, CBL, MDM2, FANCL, UBR1)
Deubiquitinating enzyme: Ataxin 3 • USP6 • CYLD
ATG3 • BIRC6 • UFC1Other Categories:- Human proteins
- Proteins
- Oncogenes
Wikimedia Foundation. 2010.