- Cyclin B1
-
G2/mitotic-specific cyclin-B1 is a protein that in humans is encoded by the CCNB1 gene.[1]
Contents
Function
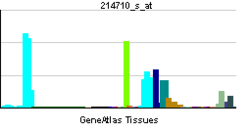
Cyclin B1 is a regulatory protein involved in mitosis. The gene product complexes with cell cycle. The different transcripts result from the use of alternate transcription initiation sites.[2]
Cyclin B1 contributes to the switch-like all or none behavior of the cell in deciding to commit to mitosis. Its activation is well regulated, and positive feedback loops ensure that once the cyclin B1-Cdk1 complex is activated it is not deactivated. Cyclin B1-Cdk1 is involved in the early events of mitosis such as chromosome condensation, nuclear envelope breakdown, and spindle pole assembly.
Once activated, cyclin B1-Cdk1 promotes several of the events of early mitosis. The active complex phosphorylates and activates 13S Condensin,[3] which helps to condense chromosomes.
Another important function of the cyclin B1-Cdk1 complex is to break down the nuclear envelope. The nuclear envelope is a membranous structure containing large protein complexes supported by a network of nuclear lamins. Phosphorylation of the lamins by cyclin B1-Cdk1 causes them to dissociate,[4] compromising the structural integrity of the nuclear envelope so that it breaks down. The destruction of the nuclear envelope is important because it allows the mitotic spindle to access the chromosomes.
Regulation
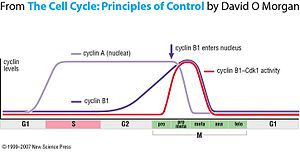
Like all cyclins, levels of cyclin B1 oscillate over the course of the cell cycle. Just prior to mitosis, a large amount of cyclin B1 is present in the cell, but it is inactive due to phosphorylation of Cdk1 by the Wee1 kinase. The complex is activated by dephosphorylation by the phosphatase Cdc25.[5] Cdc25 is always present in the cell but must be activated by phosphorylation. A possible trigger for activation is phosphorylation by cyclin A-Cdk, which functions before cyclin B1-Cdk in the cell cycle. Active Cdk1 is also capable of phosphorylating and activating Cdc25 and thus promote its own activation, resulting in a positive feedback loop. Once cyclin B1-Cdk1 is activated, it remains stably active for the rest of mitosis.
Another mechanism by which cyclin B1-Cdk1 activity is regulated is through subcellular localization. Before mitosis almost all cyclin B1 in the cell is located in the cytoplasm, but in late prophase it relocates to the nucleus. This is regulated by the phosphorylation of cyclin B1, in contrast to phosphorylation of Cdk1 regulating the activity of the complex. Phosphorylation of cyclin B1 causes it to be imported to the nucleus,[6] and phosphorylation also prevents export from the nucleus by blocking the nuclear export signal.[7] Cyclin B1 is phosphorylated by Polo kinase and Cdk1, again setting up a positive feedback loop that commits cyclin B1-Cdk1 to its fate.
At the end of mitosis, cyclin B1 is targeted for degradation by the APC through its APC localization sequence, permitting the cell to exit mitosis.
Interactions
Cyclin B1 has been shown to interact with Cdk1,[8][9][10][11] GADD45A[12][13] and RALBP1.[14]
See also
References
- ^ Sartor H, Ehlert F, Grzeschik KH, Muller R, Adolph S (Aug 1992). "Assignment of two human cell cycle genes, CDC25C and CCNB1, to 5q31 and 5q12, respectively". Genomics 13 (3): 911–2. doi:10.1016/0888-7543(92)90190-4. PMID 1386342.
- ^ "Entrez Gene: CCNB1 cyclin B1". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=891.
- ^ Kimura K, Hirano M, Kobayashi R, Hirano T (October 1998). "Phosphorylation and activation of 13S condensin by Cdc2 in vitro". Science 282 (5388): 487–90. doi:10.1126/science.282.5388.487. PMID 9774278.
- ^ Heald R, McKeon F (May 1990). "Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis". Cell 61 (4): 579–89. doi:10.1016/0092-8674(90)90470-Y. PMID 2344612.
- ^ Berry LD, Gould KL (1996). "Regulation of Cdc2 activity by phosphorylation at T14/Y15". Prog Cell Cycle Res 2: 99–105. PMID 9552387.
- ^ Hagting A, Jackman M, Simpson K, Pines J (July 1999). "Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal". Curr. Biol. 9 (13): 680–9. doi:10.1016/S0960-9822(99)80308-X. PMID 10395539.
- ^ Yang J, Song H, Walsh S, Bardes ES, Kornbluth S (February 2001). "Combinatorial control of cyclin B1 nuclear trafficking through phosphorylation at multiple sites". J. Biol. Chem. 276 (5): 3604–9. doi:10.1074/jbc.M008151200. PMID 11060306.
- ^ Shanahan, F; Seghezzi W, Parry D, Mahony D, Lees E (Feb. 1999). "Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest". Mol. Cell. Biol. 19 (2): 1460–9. PMC 116074. PMID 9891079. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=116074.
- ^ Yang, Q; Manicone A, Coursen J D, Linke S P, Nagashima M, Forgues M, Wang X W (Nov. 2000). "Identification of a functional domain in a GADD45-mediated G2/M checkpoint". J. Biol. Chem. 275 (47): 36892–8. doi:10.1074/jbc.M005319200. PMID 10973963.
- ^ Pines, J; Hunter T (Sep. 1989). "Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2". Cell 58 (5): 833–46. doi:10.1016/0092-8674(89)90936-7. PMID 2570636.
- ^ Kong, M; Barnes E A, Ollendorff V, Donoghue D J (Mar. 2000). "Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction". EMBO J. 19 (6): 1378–88. doi:10.1093/emboj/19.6.1378. PMC 305678. PMID 10716937. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=305678.
- ^ Zhan, Q; Antinore M J, Wang X W, Carrier F, Smith M L, Harris C C, Fornace A J (May. 1999). "Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45". Oncogene 18 (18): 2892–900. doi:10.1038/sj.onc.1202667. PMID 10362260.
- ^ Vairapandi, Mariappan; Balliet Arthur G, Hoffman Barbara, Liebermann Dan A (Sep. 2002). "GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress". J. Cell. Physiol. 192 (3): 327–38. doi:10.1002/jcp.10140. PMID 12124778.
- ^ Rossé, Carine; L'Hoste Sébastien, Offner Nicolas, Picard André, Camonis Jacques (Aug. 2003). "RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis". J. Biol. Chem. 278 (33): 30597–604. doi:10.1074/jbc.M302191200. PMID 12775724.
Further reading
- Kino T, Chrousos GP (2004). "Human immunodeficiency virus type-1 accessory protein Vpr: a causative agent of the AIDS-related insulin resistance/lipodystrophy syndrome?". Ann. N. Y. Acad. Sci. 1024: 153–67. doi:10.1196/annals.1321.013. PMID 15265780.
- Bailly E, Pines J, Hunter T, Bornens M (1992). "Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome.". J. Cell. Sci. 101 ( Pt 3): 529–45. PMID 1387877.
- Galaktionov K, Beach D (1992). "Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins.". Cell 67 (6): 1181–94. doi:10.1016/0092-8674(91)90294-9. PMID 1836978.
- Pines J, Hunter T (1989). "Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2.". Cell 58 (5): 833–46. doi:10.1016/0092-8674(89)90936-7. PMID 2570636.
- He J, Choe S, Walker R et al. (1995). "Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity". J. Virol. 69 (11): 6705–11. PMC 189580. PMID 7474080. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=189580.
- Re F, Braaten D, Franke EK, Luban J (1995). "Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B". J. Virol. 69 (11): 6859–64. PMC 189600. PMID 7474100. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=189600.
- Di Marzio P, Choe S, Ebright M et al. (1996). "Mutational analysis of cell cycle arrest, nuclear localization and virion packaging of human immunodeficiency virus type 1 Vpr". J. Virol. 69 (12): 7909–16. PMC 189735. PMID 7494303. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=189735.
- Jowett JB, Planelles V, Poon B et al. (1995). "The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle". J. Virol. 69 (10): 6304–13. PMC 189529. PMID 7666531. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=189529.
- Jackman M, Firth M, Pines J (1995). "Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus". EMBO J. 14 (8): 1646–54. PMC 398257. PMID 7737117. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=398257.
- Piaggio G, Farina A, Perrotti D et al. (1995). "Structure and growth-dependent regulation of the human cyclin B1 promoter". Exp. Cell Res. 216 (2): 396–402. doi:10.1006/excr.1995.1050. PMID 7843284.
- Ookata K, Hisanaga S, Bulinski JC et al. (1995). "Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics". J. Cell Biol. 128 (5): 849–62. doi:10.1083/jcb.128.5.849. PMC 2120387. PMID 7876309. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2120387.
- Pines J, Hunter T (1994). "The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B". EMBO J. 13 (16): 3772–81. PMC 395290. PMID 8070405. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=395290.
- Xiong Y, Zhang H, Beach D (1993). "Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation". Genes Dev. 7 (8): 1572–83. doi:10.1101/gad.7.8.1572. PMID 8101826.
- Zheng XF, Ruderman JV (1993). "Functional analysis of the P box, a domain in cyclin B required for the activation of Cdc25". Cell 75 (1): 155–64. doi:10.1016/S0092-8674(05)80092-3. PMID 8402895.
- O'Connor PM, Ferris DK, Pagano M et al. (1993). "G2 delay induced by nitrogen mustard in human cells affects cyclin A/cdk2 and cyclin B1/cdc2-kinase complexes differently". J. Biol. Chem. 268 (11): 8298–308. PMID 8463339.
- Sebastian B, Kakizuka A, Hunter T (1993). "Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15". Proc. Natl. Acad. Sci. U.S.A. 90 (8): 3521–4. doi:10.1073/pnas.90.8.3521. PMC 46332. PMID 8475101. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=46332.
- Kolesnitchenko V, Wahl LM, Tian H et al. (1996). "Human immunodeficiency virus 1 envelope-initiated G2-phase programmed cell death". Proc. Natl. Acad. Sci. U.S.A. 92 (25): 11889–93. doi:10.1073/pnas.92.25.11889. PMC 40508. PMID 8524869. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=40508.
- Poon RY, Jiang W, Toyoshima H, Hunter T (1996). "Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage". J. Biol. Chem. 271 (22): 13283–91. doi:10.1074/jbc.271.22.13283. PMID 8662825.
- Chen J, Saha P, Kornbluth S et al. (1996). "Cyclin-binding motifs are essential for the function of p21CIP1". Mol. Cell. Biol. 16 (9): 4673–82. PMC 231467. PMID 8756624. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=231467.
PDB gallery Cell cycle proteins Cyclin CDK CDK inhibitor P53 p63 p73 family Phases and
checkpointsOther cellular phasesB bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°) Categories:- Human proteins
- SWL/complexes with
Wikimedia Foundation. 2010.