- Protein tyrosine phosphatase
-
Protein tyrosine phosphatases (PTPs) are a group of enzymes that remove phosphate groups from phosphorylated tyrosine residues on proteins. Protein tyrosine (pTyr) phosphorylation is a common post-translational modification that can create novel recognition motifs for protein interactions and cellular localisation, affect protein stability, and regulate enzyme activity. As a consequence, maintaining an appropriate level of protein tyrosine phosphorylation is essential for many cellular functions. Tyrosine-specific protein phosphatases (PTPase; EC 3.1.3.48) catalyse the removal of a phosphate group attached to a tyrosine residue, using a cysteinyl-phosphate enzyme intermediate. These enzymes are key regulatory components in signal transduction pathways (such as the MAP kinase pathway) and cell cycle control, and are important in the control of cell growth, proliferation, differentiation and transformation[1][2].
Contents
Functions
Together with tyrosine kinases, PTPs regulate the phosphorylation state of many important signalling molecules, such as the MAP kinase family. PTPs are increasingly viewed as integral components of signal transduction cascades, despite less study and understanding compared to tyrosine kinases.
PTPs have been implicated in regulation of many cellular processes, including, but not limited to:
- Cell growth
- Cellular differentiation
- Mitotic cycles
- Oncogenic transformation
Classification
By mechanism
The PTP superfamily can be divided into four subfamilies[3][4].
Links to all 107 members of the protein tyrosine phosphatase family can be found in the template at the bottom of this article.
Class I
The class I PTPs, are the largest group of PTPs with 99 members, which can be further subdivided into
- 38 classical PTPs
- 21 receptor tyrosine phosphatase
- 17 nonreceptor-type PTPs
- 61 VH-1-like or dual-specific phosphatases (DSPs)
- 11 MAPK phosphatases (MPKs)
- 3 Slingshots
- 3 PRLs
- 4 CDC14s
- 19 atypical DSPs
- 5 Phosphatase and tensin homologs (PTENs)
- 16 Myotubularins
Dual-specificity phosphatases (dTyr and dSer/dThr) dual-specificity protein-tyrosine phosphatases. Ser/Thr and Tyr dual-specificity phosphatases are a group of enzymes with both Ser/Thr (EC 3.1.3.16) and tyrosine-specific protein phosphatase (EC 3.1.3.48) activity able to remove the serine/threonine or the tyrosine-bound phosphate group from a wide range of phosphoproteins, including a number of enzymes that have been phosphorylated under the action of a kinase. Dual-specificity protein phosphatases (DSPs) regulate mitogenic signal transduction and control the cell cycle.
LEOPARD syndrome, Noonan syndrome, and Metachondromatosis are associated with PTPN11.
Class II
LMW (low-molecular-weight) phosphatases, or acid phosphatases act on tyrosine phosphorylated proteins, low-MW aryl phosphates and natural and synthetic acyl phosphates[5][6].
The class II PTPs contain only one member, low-molecular-weight phosphotyrosine phosphatase (LMPTP).
Class III
Cdc25 phosphatases (dTyr and/or dThr)
The Class III PTPs contains three members, CDC25 A, B, and C
Class IV
pTyr-specific phosphatases
The class IV PTPs contains four members, Eya1-4.
This class is believed to have evolved separately from the other three.[7]
By location
Based on their cellular localization, PTPases are also classified as:
- Receptor-like, which are transmembrane receptors that contain PTPase domains[8]. In terms of structure, all known receptor PTPases are made up of a variable-length extracellular domain, followed by a transmembrane region and a C-terminal catalytic cytoplasmic domain. Some of the receptor PTPases contain fibronectin type III (FN-III) repeats, immunoglobulin-like domains, MAM domains, or carbonic anhydrase-like domains in their extracellular region. In general, the cytoplasmic region contains two copies of the PTPase domain. The first seems to have enzymatic activity, whereas the second is inactive.
- Non-receptor (intracellular) PTPases[9]
Common elements
All PTPases carry the highly conserved active site motif C(X)5R (PTP signature motif), employ a common catalytic mechanism, and possess a similar core structure made of a central parallel beta-sheet with flanking alpha-helices containing a beta-loop-alpha-loop that encompasses the PTP signature motif[10]. Functional diversity between PTPases is endowed by regulatory domains and subunits.
Low-molecular-weight phosphotyrosine protein phosphatase 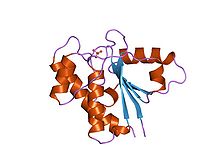
Structure of a low-molecular-weight phosphotyrosine protein phosphatase.[11] Identifiers Symbol LMWPc Pfam PF01451 InterPro IPR017867 SMART SM00226 SCOP 1phr Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Protein-tyrosine phosphatase 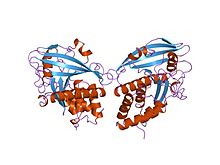
Structure of Yersinia protein tyrosine phosphatase.[12] Identifiers Symbol Y_phosphatase Pfam PF00102 Pfam clan CL0031 InterPro IPR000242 SMART SM00194 PROSITE PS50055 SCOP 1ypt Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Dual-specificity phosphatase, catalytic domain 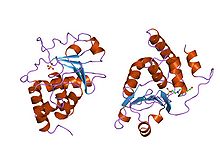
Structure of the dual-specificity protein phosphatase VHR.[13] Identifiers Symbol DSPc Pfam PF00782 Pfam clan CL0031 InterPro IPR000340 PROSITE PDOC00323 SCOP 1vhr Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Protein-tyrosine phosphatase, SIW14-like 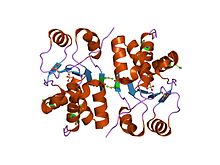
Structure of a putative phosphoprotein phosphatase from Arabidopsis thaliana.[14] Identifiers Symbol Y_phosphatase2 Pfam PF03162 Pfam clan CL0031 InterPro IPR004861 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Protein-tyrosine phosphatase-like, PTPLA Identifiers Symbol PTPLA Pfam PF04387 InterPro IPR007482 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Expression pattern
Individual PTPs may be expressed by all cell types, or their expression may be strictly tissue-specific. Most cells express 30% to 60% of all the PTPs, however hematopoietic and neuronal cells express a higher number of PTPs in comparison to other cell types. T cells and B cells of hematopoietic origin express around 60 to 70 different PTPs. The expression of several PTPS is restricted to hematopoietic cells, for example, LYP, SHP1, CD45, and HePTP.[15]
References
- ^ Dixon JE, Denu JM (1998). "Protein tyrosine phosphatases: mechanisms of catalysis and regulation". Curr Opin Chem Biol 2 (5): -. PMID 9818190.
- ^ Paul S, Lombroso PJ (2003). "Receptor and nonreceptor protein tyrosine phosphatases in the nervous system". Cell. Mol. Life Sci. 60 (11): -. doi:10.1007/s00018-003-3123-7. PMID 14625689.
- ^ Sun JP, Zhang ZY, Wang WQ (2003). "An overview of the protein tyrosine phosphatase superfamily". Curr Top Med Chem 3 (7): -. PMID 12678841.
- ^ Alonso A, Sasin J, et al. (2004). "Protein tyrosine phosphatases in the human genome". Cell 117 (6): 699–711. doi:10.1016/j.cell.2004.05.018. PMID 15186772.
- ^ Wo YY, Shabanowitz J, Hunt DF, Davis JP, Mitchell GL, Van Etten RL, McCormack AL (1992). "Sequencing, cloning, and expression of human red cell-type acid phosphatase, a cytoplasmic phosphotyrosyl protein phosphatase". J. Biol. Chem. 267 (15): 10856–10865. PMID 1587862.
- ^ Shekels LL, Smith AJ, Bernlohr DA, Van Etten RL (1992). "Identification of the adipocyte acid phosphatase as a PAO-sensitive tyrosyl phosphatase". Protein Sci. 1 (6): 710–721. doi:10.1002/pro.5560010603. PMC 2142247. PMID 1304913. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2142247.
- ^ William C. Plaxton; Michael T. McManus (2006). Control of primary metabolism in plants. Wiley-Blackwell. pp. 130–. ISBN 9781405130967. http://books.google.com/books?id=U-8GgunwoRYC&pg=PA130. Retrieved 12 December 2010.
- ^ Knapp S, Longman E, Debreczeni JE, Eswaran J, Barr AJ (2006). "The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1". Protein Sci. 15 (6): -. doi:10.1110/ps.062128706. PMC 2242534. PMID 16672235. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2242534.
- ^ Perrimon N, Johnson MR, Perkins LA, Melnick MB (1996). "The nonreceptor protein tyrosine phosphatase corkscrew functions in multiple receptor tyrosine kinase pathways in Drosophila". Dev. Biol. 180 (1): -. doi:10.1006/dbio.1996.0285. PMID 8948575.
- ^ Barford D, Das AK, Egloff MP (1998). "The structure and mechanism of protein phosphatase s: insights into catalysis and regulation". Annu. Rev. Biophys. Biomol. Struct. 27 (1): -. doi:10.1146/annurev.biophys.27.1.133. PMID 9646865.
- ^ Su XD, Taddei N, Stefani M, Ramponi G, Nordlund P (August 1994). "The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase". Nature 370 (6490): 575–8. doi:10.1038/370575a0. PMID 8052313.
- ^ Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA (August 1994). "Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate". Nature 370 (6490): 571–5. doi:10.1038/370571a0. PMID 8052312.
- ^ Yuvaniyama J, Denu JM, Dixon JE, Saper MA (May 1996). "Crystal structure of the dual specificity protein phosphatase VHR". Science 272 (5266): 1328–31. doi:10.1126/science.272.5266.1328. PMID 8650541.
- ^ Aceti DJ, Bitto E, Yakunin AF, et al. (October 2008). "Structural and functional characterization of a novel phosphatase from the Arabidopsis thaliana gene locus At1g05000". Proteins 73 (1): 241–53. doi:10.1002/prot.22041. PMID 18433060.
- ^ Mustelin T, Vang T and Bottini N. (2005). "Protein tyrosine phosphatases and the immune response". Nat. Rev. Immunol. 5 (1): 43–57. doi:10.1038/nri1530. PMID 15630428.
External links
- PTP Summary and Relevant Publications at Monash University
- MeSH Protein-Tyrosine-Phosphatase
- EC 3.1.3.48
Esterase: protein tyrosine phosphatases (EC 3.1.3.48) Class I Classical PTPsReceptor type PTPs (PTPRA, PTPRB, PTPRC, PTPRD, PTPRE, PTPRF, PTPRG, PTPRH, PTPRJ, PTPRK, PTPRM, PTPRN, PTPRN2, PTPRO, PTPRQ, PTPRR, PTPRS, PTPRT, PTPRU, PTPRZ)
Non receptor type PTPs (PTPN1, PTPN2, PTPN3, PTPN4, PTPN5, PTPN6, PTPN7, PTPN9, PTPN11, PTPN12, PTPN13, PTPN14, PTPN18, PTPN20, PTPN21, PTPN22, PTPN23MAPK phosphatases (MKPs) (DUSP1, DUSP2, DUSP4, DUSP5, DUSP6, DUSP7, DUSP8, DUSP9, DUSP10, DUSP16, MK-STYX)
CDC14s (CDC14A, CDC14B, CDKN3, PTP9Q22)
Atypical DSPs (DUSP3, DUSP11, DUSP12, DUSP13A, DUSP13B, DUSP14, DUSP15, DUSP18, DUSP19, DUSP21, DUSP22, DUSP23, DUSP24, DUSP25, DUSP26, DUSP27, EMP2A, RNGTT, STYX)
Phosphatase and tensin homologs (PTENs) (PTEN, TPIP, TPTE, TNS, TENC1)
Myotubularins (MTM1, MTMR2, MTMR3, MTMR4, MTMR5, MTMR6, MTMR7, MTMR8, MTMR9, MTMR10, MTMR11, MTMR12, MTMR13, MTMR14, MTMR15)Class II Class III Class IV MAP see MAP kinase pathwayCalcium Intracellular calcium-sensing proteins • Calcineurin • Calcium-calmodulin-dependent protein kinaseG protein cAMP: Heterotrimeric G protein (Gs/Gi) • Adenylate cyclase • cAMP • 3',5'-cyclic-AMP phosphodiesterase • Protein kinase A
cGMP: Guanylate cyclase • cGMP • 3',5'-cyclic-GMP phosphodiesterase • Protein kinase G
Beta-gamma complex Gβ (GNB1, GNB2, GNB3, GNB4, GNB5) • Gγ (GNGT1, GNGT2, GNG2, GNG3, GNG4, GNG5, GNG7, GNG8, GNG10, GNG11, GNG12, GNG13, BSCL2)
G protein-coupled receptor kinase • AMP-activated protein kinaseCyclin Lipid Phosphoinositide phospholipase C • Phospholipase c gammaOther protein kinase Serine/threonine: Casein kinase (1, 2) • eIF-2 kinase (EIF2AK3) • Glycogen synthase kinase (GSK1, GSK2, GSK-3, GSK3A, GSK3B) • IκB kinase (CHUK, IKK2, IKBKG) • Interleukin-1 receptor-associated kinase (IRAK1, IRAK2, IRAK3, IRAK4) • Lim kinase (LIMK1, LIMK2) • p21 activated kinases (PAK1, PAK2, PAK3, PAK4) • Rho-associated protein kinase (ROCK1, ROCK2) • Ribosomal s6 kinase (RPS6KA1)
Tyrosine: ZAP70 • Focal adhesion protein-tyrosine kinase (PTK2, PTK2B) • BTK
both: Dual-specificity kinaseOther phosphoprotein phosphatase Serine/threonine: Protein phosphatase 2
Tyrosine: protein tyrosine phosphatase: Receptor-like protein tyrosine phosphatase • Sh2 domain-containing protein tyrosine phosphatase
both: Dual-specificity phosphataseApoptosis see apoptosis signaling pathwayGTP-binding protein regulators Other Activating transcription factor 6 • Signal transducing adaptor protein • I-kappa B protein • Mucin-4 • Olfactory marker protein • Phosphatidylethanolamine binding protein • EDARADD • PRKCSHThis article includes text from the public domain Pfam and InterPro IPR000106
Categories:- EC 3.1.3
Wikimedia Foundation. 2010.
