- PDK3
-
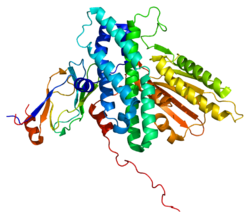
[Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3, mitochondrial is an enzyme that in humans is encoded by the PDK3 gene.[1][2] It codes for an isozyme of pyruvate dehydrogenase kinase.
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[3]
Citric_acid_cycle edit
References
- ^ Gudi R, Bowker-Kinley MM, Kedishvili NY, Zhao Y, Popov KM (Jan 1996). "Diversity of the pyruvate dehydrogenase kinase gene family in humans". J Biol Chem 270 (48): 28989–94. doi:10.1074/jbc.270.48.28989. PMID 7499431.
- ^ "Entrez Gene: PDK3 pyruvate dehydrogenase kinase, isozyme 3". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5165.
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78". http://www.wikipathways.org/index.php/Pathway:WP78.
Further reading
- Sugden MC, Holness MJ (2003). "Therapeutic potential of the mammalian pyruvate dehydrogenase kinases in the prevention of hyperglycaemia". Curr. Drug Targets Immune Endocr. Metabol. Disord. 2 (2): 151–65. doi:10.2174/1568008023340785. PMID 12476789.
- Sugden MC, Holness MJ (2003). "Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs". Am. J. Physiol. Endocrinol. Metab. 284 (5): E855–62. doi:10.1152/ajpendo.00526.2002 (inactive 2008-10-14). PMID 12676647.
- Baker JC, Yan X, Peng T, et al. (2000). "Marked differences between two isoforms of human pyruvate dehydrogenase kinase". J. Biol. Chem. 275 (21): 15773–81. doi:10.1074/jbc.M909488199. PMID 10748134.
- Kolobova E, Tuganova A, Boulatnikov I, Popov KM (2001). "Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites". Biochem. J. 358 (Pt 1): 69–77. doi:10.1042/0264-6021:3580069. PMC 1222033. PMID 11485553. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1222033.
- Korotchkina LG, Patel MS (2001). "Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase". J. Biol. Chem. 276 (40): 37223–9. doi:10.1074/jbc.M103069200. PMID 11486000.
- Tuganova A, Boulatnikov I, Popov KM (2002). "Interaction between the individual isoenzymes of pyruvate dehydrogenase kinase and the inner lipoyl-bearing domain of transacetylase component of pyruvate dehydrogenase complex". Biochem. J. 366 (Pt 1): 129–36. doi:10.1042/BJ20020301. PMC 1222743. PMID 11978179. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1222743.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=139241.
- Spriet LL, Tunstall RJ, Watt MJ, et al. (2005). "Pyruvate dehydrogenase activation and kinase expression in human skeletal muscle during fasting". J. Appl. Physiol. 96 (6): 2082–7. doi:10.1152/japplphysiol.01318.2003. PMID 14966024.
- Blackshaw S, Harpavat S, Trimarchi J, et al. (2006). "Genomic Analysis of Mouse Retinal Development". PLoS Biol. 2 (9): E247. doi:10.1371/journal.pbio.0020247. PMC 439783. PMID 15226823. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=439783.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=528928.
- Kato M, Chuang JL, Tso SC, et al. (2005). "Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex". EMBO J. 24 (10): 1763–74. doi:10.1038/sj.emboj.7600663. PMC 1142596. PMID 15861126. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1142596.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Tso SC, Kato M, Chuang JL, Chuang DT (2006). "Structural determinants for cross-talk between pyruvate dehydrogenase kinase 3 and lipoyl domain 2 of the human pyruvate dehydrogenase complex". J. Biol. Chem. 281 (37): 27197–204. doi:10.1074/jbc.M604339200. PMID 16849321.
- Devedjiev Y, Steussy CN, Vassylyev DG (2007). "Crystal Structure of an Asymmetric Complex of Pyruvate Dehydrogenase Kinase 3 with Lipoyl Domain 2 and its Biological Implications". J. Mol. Biol. 370 (3): 407–16. doi:10.1016/j.jmb.2007.04.083. PMC 1994203. PMID 17532006. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1994203.
- Degenhardt T, Saramäki A, Malinen M, et al. (2007). "Three members of the human pyruvate dehydrogenase kinase gene family are direct targets of the peroxisome proliferator-activated receptor beta/delta". J. Mol. Biol. 372 (2): 341–55. doi:10.1016/j.jmb.2007.06.091. PMID 17669420.
- Kato M, Li J, Chuang JL, Chuang DT (2007). "Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol". Structure 15 (8): 992–1004. doi:10.1016/j.str.2007.07.001. PMC 2871385. PMID 17683942. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2871385.
PDB gallery Kinases: Serine/threonine-specific protein kinases (EC 2.7.11-12) Serine/threonine-specific protein kinases (EC 2.7.11.1-EC 2.7.11.20) Non-specific serine/threonine protein kinases (EC 2.7.11.1)Pyruvate dehydrogenase kinase (EC 2.7.11.2)Dephospho-(reductase kinase) kinase (EC 2.7.11.3)(isocitrate dehydrogenase (NADP+)) kinase (EC 2.7.11.5)(tyrosine 3-monooxygenase) kinase (EC 2.7.11.6)Myosin-heavy-chain kinase (EC 2.7.11.7)Fas-activated serine/threonine kinase (EC 2.7.11.8)Goodpasture-antigen-binding protein kinase (EC 2.7.11.9)-IκB kinase (EC 2.7.11.10)cAMP-dependent protein kinase (EC 2.7.11.11)cGMP-dependent protein kinase (EC 2.7.11.12)Protein kinase C (EC 2.7.11.13)Rhodopsin kinase (EC 2.7.11.14)Beta adrenergic receptor kinase (EC 2.7.11.15)G-protein coupled receptor kinases (EC 2.7.11.16)Ca2+/calmodulin-dependent (EC 2.7.11.17)BRSK2, CAMK1, CAMK2A, CAMK2B, CAMK2D, CAMK2G, CAMK4, MLCK, CASK, CHEK1, CHEK2, DAPK1, DAPK2, DAPK3, STK11, MAPKAPK2, MAPKAPK3, MAPKAPK5, MARK1, MARK2, MARK3, MARK4, MELK, MKNK1, MKNK2, NUAK1, NUAK2, OBSCN, PASK, PHKG1, PHKG2, PIM1, PIM2, PKD1, PRKD2, PRKD3, PSKH1, SNF1LK2, KIAA0999, STK40, SNF1LK, SNRK, SPEG, TSSK2, Kalirin, TRIB1, TRIB2, TRIB3, TRIO, Titin, DCLK1Myosin light-chain kinase (EC 2.7.11.18)MYLK, MYLK2, MYLK3, MYLK4Phosphorylase kinase (EC 2.7.11.19)Elongation factor 2 kinase (EC 2.7.11.20)Serine/threonine-specific protein kinases (EC 2.7.11.21-EC 2.7.11.30) Polo kinase (EC 2.7.11.21)Cyclin-dependent kinase (EC 2.7.11.22)(RNA-polymerase)-subunit kinase (EC 2.7.11.23)Mitogen-activated protein kinase (EC 2.7.11.24)Extracellular signal-regulated (MAPK1, MAPK3, MAPK4, MAPK6, MAPK7, MAPK12, MAPK15), C-Jun N-terminal (MAPK8, MAPK9, MAPK10), P38 mitogen-activated protein (MAPK11, MAPK13, MAPK14)MAP3K (EC 2.7.11.25)Tau-protein kinase (EC 2.7.11.26)(acetyl-CoA carboxylase) kinase (EC 2.7.11.27)-Tropomyosin kinase (EC 2.7.11.28)-Low-density-lipoprotein receptor kinase (EC 2.7.11.29)-Receptor protein serine/threonine kinase (EC 2.7.11.30)Dual-specificity kinases (EC 2.7.12) Categories:- Human proteins
- Cell signaling
- Signal transduction
- Chromosome X gene stubs
Wikimedia Foundation. 2010.