- Dihydrolipoyl transacetylase
-
dihydrolipoyllysine-residue acetyltransferase 
Atomic structure of a single subunit of dihydrolipoyl transacetylase from Azotobacter vinelandii that makes up the structural core of the pyruvate dehydrogenase multienzyme complex.[1] Identifiers EC number 2.3.1.12 CAS number 9032-29-5 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles
Dihydrolipoyl transacetylase (or dihydrolipoamide acetyltransferase) is an enzyme component of the multienzyme pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is responsible for the pyruvate decarboxylation step that links glycolysis to the citric acid cycle. This involves the transformation of pyruvate from glycolysis into acetyl-CoA which is then used in the citric acid cycle to carry out cellular respiration.There are three different enzyme components in the pyruvate dehydrogenase complex. Pyruvate dehydrogenase (EC 1.2.4.1) is responsible for the oxidation of pyruvate, dihydrolipoyl transacetylase (this enzyme; EC 2.3.1.12) transfers the acetyl group to coenzyme A (CoA), and dihydrolipoyl dehydrogenase (EC 1.8.1.4) regenerates the lipoamide. Because dihydrolipoyl transacetylase is the second of the three enzyme components participating in the reaction mechanism for conversion of pyruvate into acetyl CoA, it is sometimes referred to as E2.
In humans, dihydrolipoyl transacetylase enzymatic activity resides in the pyruvate dehydrogenase complex component E2 (PDCE2) that is encoded by the DLAT (dihydrolipoamide S-acetyltransferase) gene.[3]
Contents
Nomenclature
The systematic name of this enzyme class is acetyl-CoA:enzyme N6-(dihydrolipoyl)lysine S-acetyltransferase.
Other names in common use include:
- acetyl-CoA:dihydrolipoamide S-acetyltransferase,
- acetyl-CoA:enzyme 6-N-(dihydrolipoyl)lysine S-acetyltransferase.
- dihydrolipoamide S-acetyltransferase,
- dihydrolipoate acetyltransferase,
- dihydrolipoic transacetylase,
- dihydrolipoyl acetyltransferase,
- enzyme-dihydrolipoyllysine:acetyl-CoA S-acetyltransferase,
- lipoate acetyltransferase,
- lipoate transacetylase,
- lipoic acetyltransferase,
- lipoic acid acetyltransferase,
- lipoic transacetylase,
- lipoylacetyltransferase,
- thioltransacetylase A, and
- transacetylase X.
Structure
All dihydrolipoyl transacetylases have a unique multidomain structure consisting of (from N to C): 3 lipoyl domains, an interaction domain, and the catalytic domain (see the domain architecture at Pfam). Interestingly all the domains are connected by disordered, low complexity linker regions.
Depending on the species, multiple subunits of dihydrolipoyl transacetylase enzymes can arrange together into either a cubic or dodecahedral shape. These structure then form the catalytic core of the pyruvate dehydrogenase complex which not only catalyzes the reaction that transfers an acetyl group to CoA, but also performs a crucial structural role in creating the architecture of the overall complex.[4]
 The cubic catalytic core structure made up of 24 dihydrolipoyl transacetylase subunits.[1]
The cubic catalytic core structure made up of 24 dihydrolipoyl transacetylase subunits.[1]
 The dodecahedral catalytic core structure made up of 60 dihydrolipoyl transacetylase subunits from Geobacillus stearothermophilus.[4]
The dodecahedral catalytic core structure made up of 60 dihydrolipoyl transacetylase subunits from Geobacillus stearothermophilus.[4]
Cube
The cubic core structure, found in species such as Azotobacter vinelandii, is made up of 24 subunits total.[5][6] The catalytic domains are assembled into trimers with the active site located at the subunit interface. The topology of this trimer active site is identical to that of chloramphenicol acetyltransferase. Eight of these trimers are then arranged into a hollow truncated cube. The two main substrates, CoA and the lipoamide (Lip(SH)2), are found at two opposite entrances of a 30 Å long channel which runs between the subunits and forms the catalytic center. CoA enters from the inside of the cube, and the lipoamide enters from the outside.[7]
Dodecahedron
In many species, including bacteria such as Geobacillus stearothermophilus and Enterococcus faecalis [4] as well as mammals such as humans[8] and cows,[9] the dodecahedral core structure is made up of 60 subunits total. The subunits are arranged in sets of three, similar to the trimers in the cubic core shape, with each set making up one of the 20 dodecahedral vertices.
Mechanism
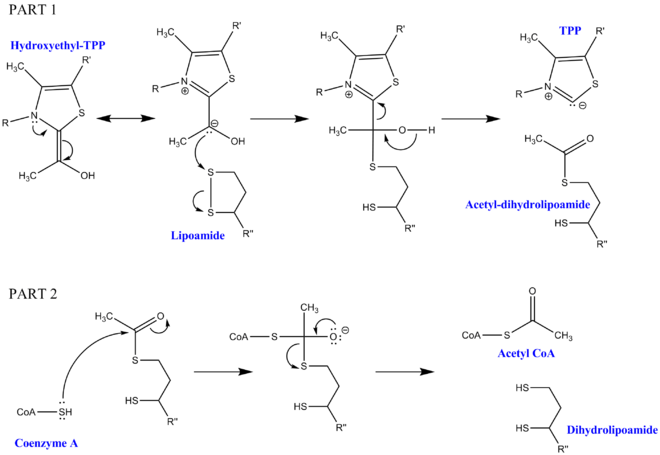
Pyruvate decarboxylation requires a few cofactors in addition to the enzymes that make up the complex. The first is thiamine pyrophosphate (TPP), which is used by pyruvate dehydrogenase to oxidize pyruvate and to form a hydroxyethyl-TPP intermediate. This intermediate is taken up by dihydrolipoyl transacetylase and reacted with a second lipoamide cofactor to generate an acetyl-dihydrolipoyl intermediate, releasing TPP in the process. This second intermediate can then be attacked by the nucleophilic sulfur attached to Coenzyme A, and the dihydrolipoamide is released. This results in the production of acetyl CoA, which is the end goal of pyruvate decarboxylation. The dihydrolipoamide is taken up by dihydrolipoyl dehydrogenase, and with the additional cofactors FAD and NAD+, regenerates the original lipoamide (with NADH as a useful side product).
Biological function and location
As mentioned above, dihydrolipoyl transacetylase participates in the pyruvate decarboxylation reaction that links glycolysis to the citric acid cycle. These metabolic processes are important for cellular respiration—the conversion of biochemical energy from nutrients into adenosine triphosphate (ATP) which can then be used to carry out numerous biological reactions within a cell. The various parts of cellular respiration take place in different parts of the cell. In eukaryotes, glycolysis occurs in the cytoplasm, pyruvate decarboxylation in the mitochondria, the citric acid cycle within the mitochondrial matrix, and oxidative phosphorylation via the electron transport chain on the mitochondrial cristae. Thus pyruvate dehydrogenase complexes (containing the dihydrolipoyl transacetylase enzymes) are found in the mitochondria of eukaryotes (and simply in the cytosol of prokaryotes).
Disease relevance
Primary biliary cirrhosis
Primary biliary cirrhosis (PBC) is an autoimmune disease characterized by autoantibodies against mitochondrial and nuclear antigens. These are called anti-mitochondrial antibodies (AMA) and anti-nuclear antibodies (ANA), respectively. These antibodies are detectable in the sera of PBC patients and vary greatly with regards to epitope specificity from patient to patient. Of the mitochondrial antigens that can generate autoantibodiy reactivity in PBC patients, the E2 subunit of the pyruvate dehydrogenase complex, dihydrolipoyl transacetylase, is the most common epitope (other antigens include enzymes of the 2-oxoacid dehydrogenase complexes as well as the other enzymes of the pyruvate dehydrogenase complexes).[10] Recent evidence has suggested that peptides within the catalytic site may present the immunodominant epitopes recognized by the anti-PDC-E2 antibodies in PBC patients.[11] There is also evidence of anti-PDC-E2 antibodies in autoimmune hepatitis (AIH) patients.[12]
Pyruvate dehydrogenase deficiency
Pyruvate dehydrogenase deficiency (PDH) is a genetic disease resulting in lactic acidosis as well as neurological dysfunction in infancy and early childhood. Typically PDH is the result of a mutation in the X-linked gene for the E1 subunit of the pyruvate dehydrogenase complex. However, there have been a few rare cases in which a patient with PDH actually has a mutation in the autosomal gene for the E2 subunit instead. These patients have been reported to have much less severe symptoms, with the most prominent disease manifestation being episodic dystonia, though both hypotonia and ataxia were also present.[13]
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[14]
Citric_acid_cycle edit
References
- ^ a b PDB 1EAF; Mattevi A, Obmolova G, Kalk KH, Teplyakov A, Hol WG (April 1993). "Crystallographic analysis of substrate binding and catalysis in dihydrolipoyl transacetylase (E2p)". Biochemistry 32 (15): 3887–901. doi:10.1021/bi00066a007. PMID 8471601.
- ^ PDB 1Y8O; Kato M, Chuang JL, Tso SC, Wynn RM, Chuang DT (May 2005). "Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex". EMBO J. 24 (10): 1763–74. doi:10.1038/sj.emboj.7600663. PMC 1142596. PMID 15861126. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1142596.
- ^ Leung PS, Watanabe Y, Munoz S, Teuber SS, Patel MS, Korenberg JR, Hara P, Coppel R, Gershwin ME (1993). "Chromosome localization and RFLP analysis of PDC-E2: the major autoantigen of primary biliary cirrhosis". Autoimmunity 14 (4): 335–40. doi:10.3109/08916939309079237. PMID 8102256.
- ^ a b c Izard T, Aevarsson A, Allen MD, et al. (February 1999). "Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes". Proceedings of the National Academy of Sciences of the United States of America 96 (4): 1240–5. Bibcode 1999PNAS...96.1240I. doi:10.1073/pnas.96.4.1240. PMC 15447. PMID 9990008. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=9990008.
- ^ de Kok A, Hengeveld AF, Martin A, Westphal AH (June 1998). "The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria". Biochimica et Biophysica Acta 1385 (2): 353–66. PMID 9655933.
- ^ Hanemaaijer R, Westphal AH, Van Der Heiden T, De Kok A, Veeger C (February 1989). "The quaternary structure of the dihydrolipoyl transacetylase component of the pyruvate dehydrogenase complex from Azotobacter vinelandii. A reconsideration". European Journal of Biochemistry / FEBS 179 (2): 287–92. doi:10.1111/j.1432-1033.1989.tb14553.x. PMID 2917567. http://www3.interscience.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0014-2956&date=1989&volume=179&issue=2&spage=287.
- ^ Mattevi A, Obmolova G, Schulze E, et al. (March 1992). "Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex". Science 255 (5051): 1544–50. Bibcode 1992Sci...255.1544M. doi:10.1126/science.1549782. PMID 1549782. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=1549782.
- ^ Brautigam CA, Wynn RM, Chuang JL, Chuang DT (May 2009). "Subunit and catalytic component stoichiometries of an in vitro reconstituted human pyruvate dehydrogenase complex". The Journal of Biological Chemistry 284 (19): 13086–98. doi:10.1074/jbc.M806563200. PMC 2676041. PMID 19240034. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=19240034.
- ^ Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ, Stoops JK (December 2001). "The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes". Proceedings of the National Academy of Sciences of the United States of America 98 (26): 14802–7. Bibcode 2001PNAS...9814802Z. doi:10.1073/pnas.011597698. PMC 64939. PMID 11752427. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=11752427.
- ^ Mackay IR, Whittingham S, Fida S, et al. (April 2000). "The peculiar autoimmunity of primary biliary cirrhosis". Immunological Reviews 174: 226–37. doi:10.1034/j.1600-0528.2002.017410.x. PMID 10807519. http://www3.interscience.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0105-2896&date=2000&volume=174&spage=226.
- ^ Braun S, Berg C, Buck S, Gregor M, Klein R (February 2010). "Catalytic domain of PDC-E2 contains epitopes recognized by antimitochondrial antibodies in primary biliary cirrhosis". World Journal of Gastroenterology : WJG 16 (8): 973–81. doi:10.3748/wjg.v16.i8.973. PMC 2828602. PMID 20180236. http://www.wjgnet.com/1007-9327/16/973.asp.
- ^ O'Brien C, Joshi S, Feld JJ, Guindi M, Dienes HP, Heathcote EJ (August 2008). "Long-term follow-up of antimitochondrial antibody-positive autoimmune hepatitis". Hepatology (Baltimore, Md.) 48 (2): 550–6. doi:10.1002/hep.22380. PMID 18666262.
- ^ Head RA, Brown RM, Zolkipli Z, et al. (August 2005). "Clinical and genetic spectrum of pyruvate dehydrogenase deficiency: dihydrolipoamide acetyltransferase (E2) deficiency". Annals of Neurology 58 (2): 234–41. doi:10.1002/ana.20550. PMID 16049940.
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78". http://www.wikipathways.org/index.php/Pathway:WP78.
Further reading
- Mattevi A, Obmolova G, Kalk KH, Teplyakov A, Hol WG (April 1993). "Crystallographic analysis of substrate binding and catalysis in dihydrolipoyl transacetylase (E2p)". Biochemistry 32 (15): 3887–901. doi:10.1021/bi00066a007. PMID 8471601.
- Brady RO, Stadtman ER (December 1954). "Enzymatic thioltransacetylation". J. Biol. Chem. 211 (2): 621–9. PMID 13221570.
- Gunsalus IC, Barton LS and Gruber W (1956). "Biosynthesis and structure of lipoic acid derivatives". J. Am. Chem. Soc. 78: 1763–1766. doi:10.1021/ja01589a079.
- Perham RN (2000). "Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions". Annu. Rev. Biochem. 69: 961–1004. doi:10.1146/annurev.biochem.69.1.961. PMID 10966480.
- Howard MJ, Fuller C, Broadhurst RW, et al. (1998). "Three-dimensional structure of the major autoantigen in primary biliary cirrhosis.". Gastroenterology 115 (1): 139–46. doi:10.1016/S0016-5085(98)70375-0. PMID 9649469.
- Matsumura S, Kita H, He XS, et al. (2002). "Comprehensive mapping of HLA-A0201-restricted CD8 T-cell epitopes on PDC-E2 in primary biliary cirrhosis.". Hepatology 36 (5): 1125–34. doi:10.1053/jhep.2002.36161. PMID 12395322.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library.". Gene 200 (1-2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Korotchkina LG, Patel MS (2008). "Binding of pyruvate dehydrogenase to the core of the human pyruvate dehydrogenase complex.". FEBS Lett. 582 (3): 468–72. doi:10.1016/j.febslet.2007.12.041. PMC 2262399. PMID 18206651. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2262399.
- Head RA, Brown RM, Zolkipli Z, et al. (2005). "Clinical and genetic spectrum of pyruvate dehydrogenase deficiency: dihydrolipoamide acetyltransferase (E2) deficiency.". Ann. Neurol. 58 (2): 234–41. doi:10.1002/ana.20550. PMID 16049940.
- Bogdanos DP, Pares A, Baum H, et al. (2004). "Disease-specific cross-reactivity between mimicking peptides of heat shock protein of Mycobacterium gordonae and dominant epitope of E2 subunit of pyruvate dehydrogenase is common in Spanish but not British patients with primary biliary cirrhosis.". J. Autoimmun. 22 (4): 353–62. doi:10.1016/j.jaut.2004.03.002. PMID 15120760.
- Lleo A, Selmi C, Invernizzi P, et al. (2009). "Apotopes and the biliary specificity of primary biliary cirrhosis.". Hepatology 49 (3): 871–9. doi:10.1002/hep.22736. PMC 2665925. PMID 19185000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2665925.
- Bellucci R, Oertelt S, Gallagher M, et al. (2007). "Differential epitope mapping of antibodies to PDC-E2 in patients with hematologic malignancies after allogeneic hematopoietic stem cell transplantation and primary biliary cirrhosis.". Blood 109 (5): 2001–7. doi:10.1182/blood-2006-06-030304. PMC 1801041. PMID 17068145. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1801041.
- Hiromasa Y, Roche TE (2003). "Facilitated interaction between the pyruvate dehydrogenase kinase isoform 2 and the dihydrolipoyl acetyltransferase.". J. Biol. Chem. 278 (36): 33681–93. doi:10.1074/jbc.M212733200. PMID 12816949.
- Trynka G, Zhernakova A, Romanos J, et al. (2009). "Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling.". Gut 58 (8): 1078–83. doi:10.1136/gut.2008.169052. PMID 19240061.
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE (2004). "Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components.". J. Biol. Chem. 279 (8): 6921–33. doi:10.1074/jbc.M308172200. PMID 14638692.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs.". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Cori CF (1981). "The glucose-lactic acid cycle and gluconeogenesis.". Curr. Top. Cell. Regul. 18: 377–87. PMID 7273846.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides.". Gene 138 (1-2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Tuganova A, Boulatnikov I, Popov KM (2002). "Interaction between the individual isoenzymes of pyruvate dehydrogenase kinase and the inner lipoyl-bearing domain of transacetylase component of pyruvate dehydrogenase complex.". Biochem. J. 366 (Pt 1): 129–36. doi:10.1042/BJ20020301. PMC 1222743. PMID 11978179. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1222743.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode 2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=139241.
External links
PDB gallery 1y8o: Crystal structure of the PDK3-L2 complex2pnr: Crystal Structure of the asymmetric Pdk3-l2 Complex1y8p: Crystal structure of the PDK3-L2 complex1fyc: INNER LIPOYL DOMAIN FROM HUMAN PYRUVATE DEHYDROGENASE (PDH) COMPLEX, NMR, 1 STRUCTURE1y8n: Crystal structure of the PDK3-L2 complex2dne: Solution Structure of RSGI RUH-058, a lipoyl domain of human 2-oxoacid dehydrogenase2q8i: Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicolTransferases: acyltransferases (EC 2.3) 2.3.1: other than amino-acyl groups acetyltransferases: Acetyl-Coenzyme A acetyltransferase - N-Acetylglutamate synthase - Choline acetyltransferase - Dihydrolipoyl transacetylase - Acetyl-CoA C-acyltransferase - Beta-galactoside transacetylase - Chloramphenicol acetyltransferase - N-acetyltransferase (Serotonin N-acetyl transferase, HGSNAT, ARD1A) - Histone acetyltransferase (P300/CBP, NAT2)
palmitoyltransferases: Carnitine O-palmitoyltransferase (CPT1, CPT2) - Serine C-palmitoyltransferase (SPTLC1, SPTLC2)
other: Acyltransferase like 2 - Aminolevulinic acid synthase - Beta-ketoacyl-ACP synthase - Glyceronephosphate O-acyltransferase - Lecithin-cholesterol acyltransferase
Glycerol-3-phosphate O-acyltransferase - 1-acylglycerol-3-phosphate O-acyltransferase - 2-acylglycerol-3-phosphate O-acyltransferase - ABHD52.3.2: Aminoacyltransferases 2.3.3: converted into alkyl on transfer Photosynthesis Dehydrogenase Other CAD (Carbamoyl phosphate synthase II, Aspartate carbamoyltransferase, Dihydroorotase) · Cholesterol side-chain cleavage enzyme · Cytochrome b6f complex · Electron transport chain · Fatty acid synthetase complex · Glycine decarboxylase complex · Mitochondrial trifunctional protein (HADHA, HADHB) · Phosphoenolpyruvate sugar phosphotransferase system · Polyketide synthase · Sucrase-isomaltase complex · Tryptophan synthaseGlycolysis Metabolic Pathway Glucose Hexokinase Glucose 6-phosphate Glucose-6-phosphate isomerase Fructose 6-phosphate phosphofructokinase-1 Fructose 1,6-bisphosphate Fructose bisphosphate aldolase 
ATP ADP 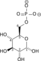
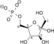
ATP ADP 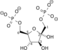


Dihydroxyacetone phosphate Glyceraldehyde 3-phosphate Triosephosphate isomerase Glyceraldehyde 3-phosphate Glyceraldehyde-3-phosphate dehydrogenase 1,3-Bisphosphoglycerate 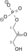
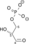

NAD+ + Pi NADH + H+ 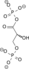
+ 
2 
2 Phosphoglycerate kinase 3-Phosphoglycerate Phosphoglycerate mutase 2-Phosphoglycerate Phosphopyruvate hydratase(Enolase) Phosphoenolpyruvate Pyruvate kinase Pyruvate ADP ATP 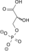
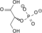
H2O 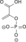
ADP ATP 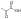
2 
2 
2 
2 Categories:- Human proteins
- Glycolysis
Wikimedia Foundation. 2010.