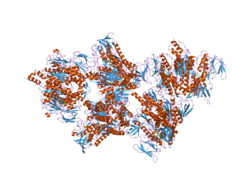- Dihydrolipoamide dehydrogenase
-
dihydrolipoyl dehydrogenase Identifiers EC number 1.8.1.4 CAS number 9001-18-7 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the DLD gene.[1][2][3][4] DLD is a flavoprotein enzyme that degrades lipoamide, and produces dihydrolipoamide.
Contents
Function
This gene encodes the L protein of the mitochondrial glycine cleavage system. The L protein, also named dihydrolipoamide dehydrogenase, is also a component of the pyruvate dehydrogenase complex, the alpha-ketoglutarate dehydrogenase complex, and the branched-chain alpha-keto acide dehydrogenase complex.[1]
Clinical significance
Mutations in this gene have been identified in patients with E3-deficient maple syrup urine disease and lipoamide dehydrogenase deficiency.[1]
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[5]
Citric_acid_cycle edit
See also
References
- ^ a b c "Entrez Gene: dihydrolipoamide dehydrogenase". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1738.
- ^ Otulakowski G, Robinson BH (December 1987). "Isolation and sequence determination of cDNA clones for porcine and human lipoamide dehydrogenase. Homology to other disulfide oxidoreductases". J. Biol. Chem. 262 (36): 17313–8. PMID 3693355.
- ^ Pons G, Raefsky-Estrin C, Carothers DJ, Pepin RA, Javed AA, Jesse BW, Ganapathi MK, Samols D, Patel MS (March 1988). "Cloning and cDNA sequence of the dihydrolipoamide dehydrogenase component human alpha-ketoacid dehydrogenase complexes". Proc. Natl. Acad. Sci. U.S.A. 85 (5): 1422–6. doi:10.1073/pnas.85.5.1422. PMC 279783. PMID 3278312. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=279783.
- ^ Scherer SW, Otulakowski G, Robinson BH, Tsui LC (1991). "Localization of the human dihydrolipoamide dehydrogenase gene (DLD) to 7q31----q32". Cytogenet. Cell Genet. 56 (3-4): 176–7. PMID 2055113.
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78". http://www.wikipathways.org/index.php/Pathway:WP78.
Further reading
- Silverberg MS, Cho JH, Rioux JD, et al. (2009). "Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study.". Nat. Genet. 41 (2): 216–20. doi:10.1038/ng.275. PMC 2652837. PMID 19122664. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2652837.
- Scherer SW, Cheung J, MacDonald JR, et al. (2003). "Human chromosome 7: DNA sequence and biology.". Science 300 (5620): 767–72. doi:10.1126/science.1083423. PMC 2882961. PMID 12690205. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2882961.
- Brautigam CA, Chuang JL, Tomchick DR, et al. (2005). "Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations.". J. Mol. Biol. 350 (3): 543–52. doi:10.1016/j.jmb.2005.05.014. PMID 15946682.
- , Barrett JC, Lee JC, et al. (2009). "Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region.". Nat. Genet. 41 (12): 1330–4. doi:10.1038/ng.483. PMID 19915572.
- Reed LJ, Hackert ML (1990). "Structure-function relationships in dihydrolipoamide acyltransferases.". J. Biol. Chem. 265 (16): 8971–4. PMID 2188967.
- Ciszak EM, Makal A, Hong YS, et al. (2006). "How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex.". J. Biol. Chem. 281 (1): 648–55. doi:10.1074/jbc.M507850200. PMID 16263718.
- Asano K, Matsushita T, Umeno J, et al. (2009). "A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population.". Nat. Genet. 41 (12): 1325–9. doi:10.1038/ng.482. PMID 19915573.
- Odièvre MH, Chretien D, Munnich A, et al. (2005). "A novel mutation in the dihydrolipoamide dehydrogenase E3 subunit gene (DLD) resulting in an atypical form of alpha-ketoglutarate dehydrogenase deficiency.". Hum. Mutat. 25 (3): 323–4. doi:10.1002/humu.9319. PMID 15712224.
- Brautigam CA, Wynn RM, Chuang JL, et al. (2006). "Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex.". Structure 14 (3): 611–21. doi:10.1016/j.str.2006.01.001. PMID 16442803.
- Kim H (2006). "Activity of human dihydrolipoamide dehydrogenase is largely reduced by mutation at isoleucine-51 to alanine.". J. Biochem. Mol. Biol. 39 (2): 223–7. PMID 16584639.
- Sugden MC, Holness MJ (2003). "Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs.". Am. J. Physiol. Endocrinol. Metab. 284 (5): E855-62. doi:10.1152/ajpendo.00526.2002. PMID 12676647.
- Wang YC, Wang ST, Li C, et al. (2008). "The role of amino acids T148 and R281 in human dihydrolipoamide dehydrogenase.". J. Biomed. Sci. 15 (1): 37–46. doi:10.1007/s11373-007-9208-9. PMID 17960497.
- Brown AM, Gordon D, Lee H, et al. (2004). "Association of the dihydrolipoamide dehydrogenase gene with Alzheimer's disease in an Ashkenazi Jewish population.". Am. J. Med. Genet. B Neuropsychiatr. Genet. 131B (1): 60–6. doi:10.1002/ajmg.b.30008. PMID 15389771.
- Babady NE, Pang YP, Elpeleg O, Isaya G (2007). "Cryptic proteolytic activity of dihydrolipoamide dehydrogenase.". Proc. Natl. Acad. Sci. U.S.A. 104 (15): 6158–63. doi:10.1073/pnas.0610618104. PMID 17404228.
- Wang YC, Wang ST, Li C, et al. (2007). "The role of N286 and D320 in the reaction mechanism of human dihydrolipoamide dehydrogenase (E3) center domain.". J. Biomed. Sci. 14 (2): 203–10. doi:10.1007/s11373-006-9136-0. PMID 17171578.
- Foster LJ, Rudich A, Talior I, et al. (2006). "Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC).". J. Proteome Res. 5 (1): 64–75. doi:10.1021/pr0502626. PMID 16396496.
- Kim H (2005). "Asparagine-473 residue is important to the efficient function of human dihydrolipoamide dehydrogenase.". J. Biochem. Mol. Biol. 38 (2): 248–52. PMID 15826505.
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE (2004). "Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components.". J. Biol. Chem. 279 (8): 6921–33. doi:10.1074/jbc.M308172200. PMID 14638692.
- Wynn RM, Kato M, Machius M, et al. (2004). "Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation.". Structure 12 (12): 2185–96. doi:10.1016/j.str.2004.09.013. PMID 15576032.
- Martins-de-Souza D, Gattaz WF, Schmitt A, et al. (2009). "Proteome analysis of schizophrenia patients Wernicke's area reveals an energy metabolism dysregulation.". BMC Psychiatry 9: 17. doi:10.1186/1471-244X-9-17. PMC 2684104. PMID 19405953. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2684104.
External links
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
PDB gallery 1zy8: The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex.2f5z: Crystal Structure of Human Dihydrolipoamide Dehydrogenase (E3) Complexed to the E3-Binding Domain of Human E3-Binding Protein1zmc: Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NAD+1zmd: Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NADHOxidoreductases: sulfur oxidoreductases (EC 1.8) 1.8.1: NAD or NADP Dihydrolipoamide dehydrogenase - Glutathione reductase - Thioredoxin reductase1.8.2: cytochrome 1.8.3: oxygen 1.8.4: disulfide 1.8.5: quinone 1.8.98: Other, known 1.8.99: Other Aldehyde/oxo oxidoreductases (EC 1.2) 1.2.1: NAD or NADP 1.2.2: cytochrome 1.2.3: oxygen 1.2.4: disulfide 1.2.7: iron-sulfur protein Acetolactate synthase - Acyl CoA dehydrogenase - Apoptosis-inducing factor - Butyryl CoA dehydrogenase - Cryptochrome - Cytochrome b5 reductase - Dihydrolipoamide dehydrogenase - Flavodoxin - Methemoglobin reductase - Methylenetetrahydrofolate reductase - NADH dehydrogenase - NADPH oxidase - Nitrate reductase - Sarcosine oxidase - Thioredoxin reductasePhotosynthesis Dehydrogenase Pyruvate dehydrogenase complex (E1, E2, E3) · Oxoglutarate dehydrogenase (OGDH, DLST, DLD) · Branched-chain alpha-keto acid dehydrogenase complex (BCKDHA, BCKDHB, DBT, DLD)Other CAD (Carbamoyl phosphate synthase II, Aspartate carbamoyltransferase, Dihydroorotase) · Cholesterol side-chain cleavage enzyme · Cytochrome b6f complex · Electron transport chain · Fatty acid synthetase complex · Glycine decarboxylase complex · Mitochondrial trifunctional protein (HADHA, HADHB) · Phosphoenolpyruvate sugar phosphotransferase system · Polyketide synthase · Sucrase-isomaltase complex · Tryptophan synthaseGlycolysis Metabolic Pathway Glucose Hexokinase Glucose 6-phosphate Glucose-6-phosphate isomerase Fructose 6-phosphate phosphofructokinase-1 Fructose 1,6-bisphosphate Fructose bisphosphate aldolase 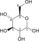
ATP ADP 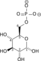
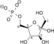
ATP ADP 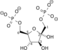


Dihydroxyacetone phosphate Glyceraldehyde 3-phosphate Triosephosphate isomerase Glyceraldehyde 3-phosphate Glyceraldehyde-3-phosphate dehydrogenase 1,3-Bisphosphoglycerate 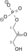
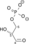

NAD+ + Pi NADH + H+ 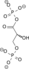
+ 
2 
2 Phosphoglycerate kinase 3-Phosphoglycerate Phosphoglycerate mutase 2-Phosphoglycerate Phosphopyruvate hydratase(Enolase) Phosphoenolpyruvate Pyruvate kinase Pyruvate ADP ATP 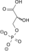
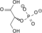
H2O 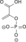
ADP ATP 
2 
2 
2 
2 This oxidoreductase article is a stub. You can help Wikipedia by expanding it.