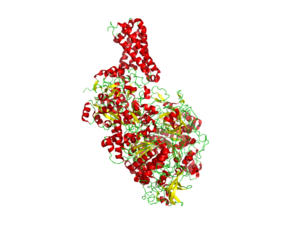
- Nitrate reductase
-
Nitrate reductases are molybdoenzymes that reduce nitrate (NO−
3) to nitrite (NO−
2).- Eukaryotic nitrate reductases are part of the sulfite oxidase family of molybdoenzymes.
- Prokaryotic nitrate reductases belong to the DMSO reductase family of molybdoenzymes and have been classified into three groups, assimilatory nitrate reductases (Nas), respiratory nitrate reductase (Nar), and periplasmic nitrate reductases (Nap). The active site of these enzymes is a Mo ion that is bound to the four thiolate functions of two pterin molecules. The coordination sphere of the Mo is completed by one amino-acid side chain and oxygen and/or sulfur ligands. The exact environment of the Mo ion in certain of these enzymes (oxygen versus sulfur as a sixth molybdenum ligand) is still debated. The Mo is covalently attached to the protein by a cysteine ligand in Nap, and an aspartate in Nar.[1]
Catalytic mechanism
Nitrate molecule binds to the active site with the Mo ion in the +6 oxidation state. Electron transfer to the active site occurs only in the proton-electron transfer stage, where the MoV species plays an important role in catalysis. The presence of the sulfur atom in the molybdenum coordination sphere creates a pseudo-dithiolene ligand that protects it from any direct attack from the solvent. Upon the nitrate binding there is a conformational rearrangement of this ring that allows the direct contact of the nitrate with MoVI ion. This rearrangement is stabilized by the conserved methionines Met141 and Met308. The reduction of nitrate into nitrite occurs in the second step of the mechanism where the two dimethyl-dithiolene ligands have a key role in spreading the excess of negative charge near the Mo atom to make it available for the chemical reaction. The reaction involves the oxidation of the sulfur atoms and not of the molybdenum as previously suggested. The mechanism involves a molybdenum and sulfur-based redox chemistry instead of the currently accepted redox chemistry based only on the Mo ion. The second part of the mechanism involves two protonation steps that are promoted by the presence of MoV species. MoVI intermediates might also be present in this stage depending on the availability of protons and electrons. Once the water molecule is generated only the MoVI species allow water molecule dissociation, and, the concomitant enzymatic turnover. [2]
References
- ^ Tavares, P; Tavares, P., Pereira, A. S., Moura, J J G, Moura, I (2006). "Metalloenzymes of the denitrification pathway". Journal of Inorganic Biochemistry 100 (12): 2087–2100. doi:10.1016/j.jinorgbio.2006.09.003. PMID 17070915.
- ^ N. M. F. S. A. Cerqueira, P. J. Gonzalez, C. D. Brondino, M. J. Romão, C. C. Romão, I. Moura, J. J. G. Moura (2009). "The effect of the sixth sulfur ligand in the catalytic mechanism of periplasmic nitrate reductase". Journal of Computational Chemistry 30 (15): 2466–2484. doi:10.1002/jcc.21280.
External links
This enzyme-related article is a stub. You can help Wikipedia by expanding it.