- Chloramphenicol acetyltransferase
-
Chloramphenicol acetyltransferase 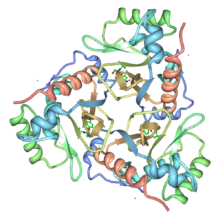
Ribbon diagram of the chloramphenicol acetyltransferase trimer with chloramphenicol bound. From PDB 3CLA. Identifiers Symbol CAT Pfam PF00302 InterPro IPR001707 PROSITE PDOC00093 SCOP 3cla Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Chloramphenicol acetyltransferase (or CAT) is a bacterial enzyme (EC 2.3.1.28)[1] that detoxifies the antibiotic chloramphenicol and is responsible for chloramphenicol resistance in bacteria.[2] This enzyme covalently attaches an acetyl group from acetyl-CoA to chloramphenicol, which prevents chloramphenicol from binding to ribosomes. A histidine residue, located in the C-terminal section of the enzyme, plays a central role in its catalytic mechanism.
The crystal structure of the type III enzyme from Escherichia coli with chloramphenicol bound has been determined. CAT is a trimer of identical subunits (monomer Mr 25,000) and the trimeric structure is stabilised by a number of hydrogen bonds, some of which result in the extension of a beta-sheet across the subunit interface. Chloramphenicol binds in a deep pocket located at the boundary between adjacent subunits of the trimer, such that the majority of residues forming the binding pocket belong to one subunit while the catalytically essential histidine belongs to the adjacent subunit. His195 is appropriately positioned to act as a general base catalyst in the reaction, and the required tautomeric stabilisation is provided by an unusual interaction with a main-chain carbonyl oxygen[3].
Application
CAT is used as a reporter system to measure the level of a promoter or its tissue-specific expression. The CAT assay involves monitoring acetylation of radioactive chloramphenicol on a TCA plate; we determine CAT activity by looking for one or both of the two acetylated forms of chloramphenicol, which have a significantly increased migration rate as compared to the unacetylated form.
References
- ^ Engel J, Prockop DJ (1991). "The zipper-like folding of collagen triple helices and the effects of mutations that disrupt the zipper". Annu. Rev. Biophys. Biophys. Chem. 20 (1): 137–152. doi:10.1146/annurev.bb.20.060191.001033. PMID 1867713.
- ^ Shaw WV, Packman LC, Burleigh BD, Dell A, Morris HR, Hartley BS (1979). "Primary structure of a chloramphenicol acetyltransferase specified by R plasmids". Nature 282 (5741): 870–2. doi:10.1038/282870a0. PMID 390404.
- ^ Leslie AG (1990). "Refined crystal structure of type III chloramphenicol acetyltransferase at 1.75 A resolution". J. Mol. Biol. 213 (1): 167–186. doi:10.1016/S0022-2836(05)80129-9. PMID 2187098.
Transferases: acyltransferases (EC 2.3) 2.3.1: other than amino-acyl groups acetyltransferases: Acetyl-Coenzyme A acetyltransferase - N-Acetylglutamate synthase - Choline acetyltransferase - Dihydrolipoyl transacetylase - Acetyl-CoA C-acyltransferase - Beta-galactoside transacetylase - Chloramphenicol acetyltransferase - N-acetyltransferase (Serotonin N-acetyl transferase, HGSNAT, ARD1A) - Histone acetyltransferase (P300/CBP, NAT2)
palmitoyltransferases: Carnitine O-palmitoyltransferase (CPT1, CPT2) - Serine C-palmitoyltransferase (SPTLC1, SPTLC2)
other: Acyltransferase like 2 - Aminolevulinic acid synthase - Beta-ketoacyl-ACP synthase - Glyceronephosphate O-acyltransferase - Lecithin-cholesterol acyltransferase
Glycerol-3-phosphate O-acyltransferase - 1-acylglycerol-3-phosphate O-acyltransferase - 2-acylglycerol-3-phosphate O-acyltransferase - ABHD52.3.2: Aminoacyltransferases 2.3.3: converted into alkyl on transfer This article includes text from the public domain Pfam and InterPro IPR001707
Categories:- EC 2.3.1
- Bacterial enzymes
- Bacteria stubs
- Biochemistry stubs
Wikimedia Foundation. 2010.
