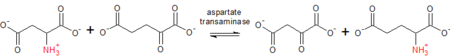- Aspartate transaminase
-
aspartate transaminase 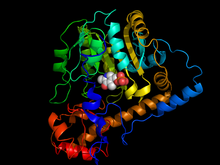
Aspartate aminotransferase from Escherichia coli bound with cofactor pyridoxal 5-phosphate.[1] Identifiers EC number 2.6.1.1 CAS number 9000-97-9 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles Aspartate transaminase (AST), also called aspartate aminotransferase (AspAT/ASAT/AAT) or serum glutamic oxaloacetic transaminase (SGOT), is a pyridoxal phosphate (PLP)-dependent transaminase enzyme (EC 2.6.1.1). AST catalyzes the reversible transfer of an α-amino group between aspartate and glutamate and, as such, is an important enzyme in amino acid metabolism. AST is found in the liver, heart, skeletal muscle, kidneys, brain, and red blood cells, and it is commonly measured clinically as a marker for liver health.
Contents
Function
Aspartate transaminase catalyzes the interconversion of aspartate and α-ketoglutarate to oxaloacetate and glutamate.
Aspartate (Asp) + α-ketoglutarate ⇌ oxaloacetate + glutamate (Glu)
As a prototypical transaminase, AST relies on PLP as a cofactor to transfer the amino group from aspartate or glutamate to the corresponding ketoacid. In the process, the cofactor shuttles between PLP and the pyridoxamine phosphate (PMP) form.[2] The amino group transfer catalyzed by this enzyme is crucial in both amino acid degradation and biosynthesis. In amino acid degradation, following the conversion of aspartate to glutamate, glutamate subsequently undergoes oxidative deamidation to form ammonium ions, which are excreted as urea. In the reverse reaction, aspartate may be synthesized from oxaloacetate, which is a key intermediate in the citric acid cycle.[3]
Isozymes
Two isoenzymes are present in a wide variety of eukaryotes. In humans:
- GOT1/cAST, the cytosolic isoenzyme derives mainly from red blood cells and heart.
- GOT2/mAST, the mitochondrial isoenzyme is present predominantly in liver.
These isoenzymes are thought to have evolved from a common ancestral AST via gene duplication, and they share a sequence homology of approximately 45%.[4]
AST has also been found in a number of microorganisms, including E. coli, H. mediterranei,[5] and T. thermophilus.[6] In E. coli, the enzyme is encoded by the aspCgene and has also been shown to exhibit the activity of an aromatic-amino-acid transaminase (EC 2.6.1.57).[7]
Structure
X-ray crystallography studies have been performed to determine the structure of aspartate transaminase from various sources, including chicken mitochondria,[8] pig heart cytosol,[9] and E. coli.[10][11] Overall, the three-dimensional polypeptide structure for all species is quite similar. AST is dimeric, consisting of two identical subunits, each with approximately 400 amino acid residues and a molecular weight of approximately 45 kD.[4] Each subunit is composed of a large and a small domain, as well as a third domain consisting of the N-terminal residues 3-14; these few residues form a strand, which links and stabilizes the two subunits of the dimer. The large domain, which includes residues 48-325, binds the PLP cofactor via an aldimine linkage to the ε-amino group of Lys258. Other residues in this domain – Asp 222 and Tyr 225 – also interact with PLP via hydrogen bonding. The small domain consists of residues 15-47 and 326-410 and represents a flexible region that shifts the enzyme from an "open" to a "closed" conformation upon substrate binding.[8][11][12]
The two independent active sites are positioned near the interface between the two domains. Within each active site, a couple arginine residues are responsible for the enzyme’s specificity for dicarboxylic acid substrates: Arg386 interacts with the substrate’s proximal (α-)carboxylate group, while Arg292 complexes with the distal (side-chain) carboxylate.[8][11]
In terms of secondary structure, AST contains both α and β elements. Each domain has a central sheet of β-strands with α-helices packed on either side.
Mechanism
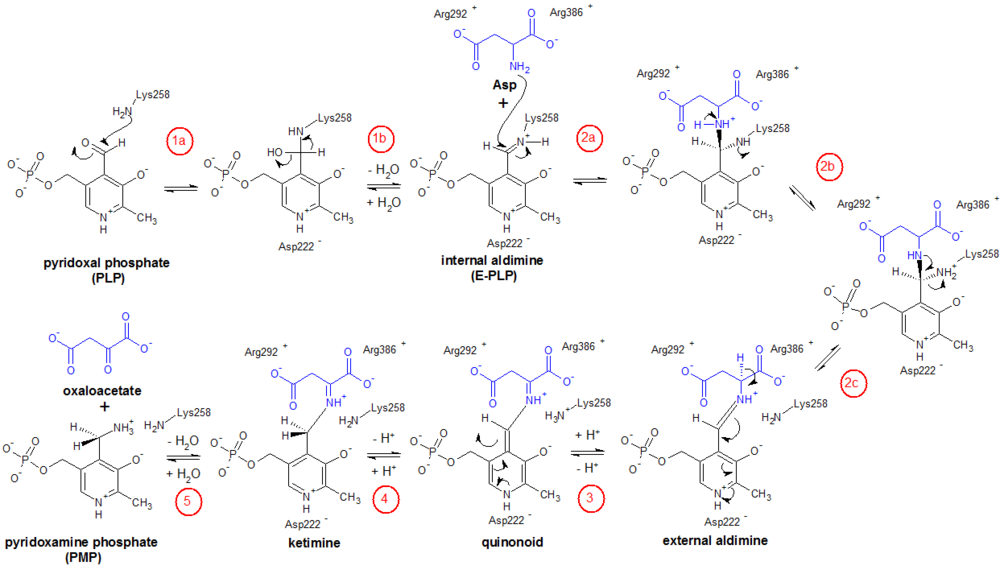
Aspartate transaminase, as with all transaminases, operates via dual substrate recognition; that is, it is able to recognize and selectively bind two amino acids (Asp and Glu) with different side-chains.[13] In either case, the transaminase reaction consists of two similar half-reactions that constitute what is referred to as a ping-pong mechanism. In the first half-reaction, amino acid 1 (e.g., L-Asp) reacts with the enzyme-PLP complex to generate ketoacid 1 (oxaloacetate) and the modified enzyme-PMP. In the second half-reaction, ketoacid 2 (α-ketoglutarate) reacts with enzyme-PMP to produce amino acid 2 (L-Glu), regenerating the original enzyme-PLP in the process. Formation of a racemic product (D-Glu) is very rare.[14]
The specific steps for the half-reaction of Enzyme-PLP + aspartate ⇌ Enzyme-PMP + oxaloacetate are as follows (see figure); the other half-reaction (not shown) proceeds in the reverse manner, with α-ketoglutarate as the substrate.[2][3]
- Internal aldimine formation: First, the ε-amino group of Lys258 forms a Schiff base linkage with the aldehyde carbon to generate an internal aldimine.
- Transaldimination: The internal aldimine then becomes an external aldimine when the ε-amino group of Lys258 is displaced by the amino group of aspartate. This transaldimination reaction occurs via a nucleophilic attack by the deprotonated amino group of Asp and proceeds through a tetrahedral intermediate. As this point, the carboxylate groups of Asp are stabilized by the guanidinium groups of the enzyme’s Arg386 and Arg 292 residues.
- Quinonoid formation: The hydrogen attached to the a-carbon of Asp is then abstracted (Lys258 is thought to be the proton acceptor) to form a quinonoid intermediate.
- Ketimine formation: The quinonoid is reprotonated, but now at the aldehyde carbon, to form the ketimine intermediate.
- Ketimine hydrolysis: Finally, the ketimine is hydrolyzed to form PMP and oxaloacetate.
This mechanism is thought to have multiple partially rate-determining steps.[15] However, it has been shown that the substrate binding step (transaldimination) drives the catalytic reaction forward.[16]
Clinical significance
AST is similar to alanine transaminase (ALT) in that both enzymes are associated with liver parenchymal cells. The difference is that ALT is found predominantly in the liver, with clinically negligible quantities found in the kidneys, heart, and skeletal muscle, while AST is found in the liver, heart (cardiac muscle), skeletal muscle, kidneys, brain, and red blood cells.[17] As a result, ALT is a more specific indicator of liver inflammation than AST, as AST may be elevated also in diseases affecting other organs, such as myocardial infarction, acute pancreatitis, acute hemolytic anemia, severe burns, acute renal disease, musculoskeletal diseases, and trauma.[18]
AST was defined as a biochemical marker for the diagnosis of acute myocardial infarction in 1954. However, the use of AST for such a diagnosis is now redundant and has been superseded by the cardiac troponins.[19]
AST (SGOT) is commonly measured clinically as a part of diagnostic liver function tests, to determine liver health.
Patient type Reference ranges[20] Female 6 - 34 IU/L Male 8 - 40 IU/L It has also been shown to be a marker for chronic alcoholism. [21]
See also
- Transaminases
- Alanine transaminase
References
- ^ PDB 1AAMAlmo SC, Smith DL, Danishefsky AT, Ringe D (March 1994). "The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli". Protein Eng. 7 (3): 405–412. doi:10.1093/protein/7.3.405. PMID 7909946.
- ^ a b Kirsch JF, Eichele G, Ford GC, Vincent MG, Jansonius JN, Gehring H et al. (1984). "Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure". J Mol Biol 174 (3): 497–525. doi:10.1016/0022-2836(84)90333-4. PMID 6143829. http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=6143829.
- ^ a b Berg, JM; Tymoczko, JL; Stryer, L (2006). Biochemistry. W.H. Freeman. pp. 656–660. ISBN 978-0716787242.
- ^ a b Hayashi H, Wada H, Yoshimura T, Esaki N, Soda K (1990). "Recent topics in pyridoxal 5'-phosphate enzyme studies". Annu Rev Biochem 59: 87–110. doi:10.1146/annurev.bi.59.070190.000511. PMID 2197992.
- ^ Muriana FJ, Alvarez-Ossorio MC, Relimpio AM (1991). "Purification and characterization of aspartate aminotransferase from the halophile archaebacterium Haloferax mediterranei". Biochem J 278 ( Pt 1): 149–54. PMC 1151461. PMID 1909112. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1151461.
- ^ Okamoto A, Kato R, Masui R, Yamagishi A, Oshima T, Kuramitsu S (1996). "An aspartate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus HB8". J Biochem 119 (1): 135–44. PMID 8907187.
- ^ Gelfand DH, Steinberg RA (1977). "Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases". J Bacteriol 130 (1): 429–40. PMC 235221. PMID 15983. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=235221.
- ^ a b c McPhalen CA, Vincent MG, Jansonius JN (1992). "X-ray structure refinement and comparison of three forms of mitochondrial aspartate aminotransferase". J Mol Biol 225 (2): 495–517. doi:10.1016/0022-2836(92)90935-D. PMID 1593633. http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=1593633.
- ^ Rhee S, Silva MM, Hyde CC, Rogers PH, Metzler CM, Metzler DE et al. (1997). "Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate". J Biol Chem 272 (28): 17293–302. doi:10.1074/jbc.272.28.17293. PMID 9211866.
- ^ Kamitori S, Hirotsu K, Higuchi T, Kondo K, Inoue K, Kuramitsu S et al. (1988). "Three-dimensional structure of aspartate aminotransferase from Escherichia coli at 2.8 A resolution". J Biochem 104 (3): 317–8. PMID 3071527.
- ^ a b c Danishefsky AT, Onnufer JJ, Petsko GA, Ringe D (1991). "Activity and structure of the active-site mutants R386Y and R386F of Escherichia coli aspartate aminotransferase". Biochemistry 30 (7): 1980–1985. doi:10.1021/bi00221a035. PMID 1993208.
- ^ McPhalen CA, Vincent MG, Picot D, Jansonius JN, Lesk AM, Chothia C (1992). "Domain closure in mitochondrial aspartate aminotransferase". J Mol Biol 227 (1): 197–213. doi:10.1016/0022-2836(92)90691-C. PMID 1522585. http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=1522585.
- ^ Hirotsu K, Goto M, Okamoto A, Miyahara I (2005). "Dual substrate recognition of aminotransferases". Chem Rec 5 (3): 160–172. doi:10.1002/tcr.20042. PMID 15889412.
- ^ Kochhar S, Christen P (1992). "Mechanism of racemization of amino acids by aspartate aminotransferase". Eur J Biochem 203 (3): 563–569. doi:10.1111/j.1432-1033.1992.tb16584.x. PMID 1735441.
- ^ Goldberg JM, Kirsch JF (1996). "The reaction catalyzed by Escherichia coli aspartate aminotransferase has multiple partially rate-determining steps, while that catalyzed by the Y225F mutant is dominated by ketimine hydrolysis". Biochemistry 35 (16): 5280–5291. doi:10.1021/bi952138d. PMID 8611515. http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=8611515.
- ^ Hayashi H, Mizuguchi H, Miyahara I, Nakajima Y, Hirotsu K, Kagamiyama H (2003). "Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis". J Biol Chem 278 (11): 9481–9488. doi:10.1074/jbc.M209235200. PMID 12488449. http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=12488449.
- ^ http://dynaweb.ebscohost.com/Detail?sid=923b5a81-7daf-46b7-bdb2-86d8649da6ef@sessionmgr13&vid=&db=dme&ss=AN+%22316452%22&sl=ll
- ^ http://www.rnceus.com/lf/lfast.html
- ^ Gaze DC (2007). "The role of existing and novel cardiac biomarkers for cardioprotection". Curr. Opin. Invest. Drugs 8 (9): 711–7. PMID 17729182.
- ^ GPnotebook > reference range (AST) Retrieved on Dec 7, 2009
- ^ Nalpas B, Vassault A, Charpin S, Lacour B, Berthelot P (1986). "Serum mitochondrial aspartate aminotransferase as a marker of chronic alcoholism: diagnostic value and interpretation in a liver unit". Hepatology 6 (4): 608–614. doi:10.1002/hep.1840060410. PMID 3732996. http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=3732996.
Further reading
- Jansonius, JN; Vincent, MG (1987). Jurnak FA and McPherson A. ed. Structural basis for catalysis by aspartate aminotransferase (Biological Macromolecules and Assemblies, Vol. 3). New York: Wiley. pp. 187–285. ISBN 0-471-85142-6.
- Kuramitsu S, Okuno S, Ogawa T, Ogawa H, Kagamiyama H (1985). "Aspartate aminotransferase of Escherichia coli: nucleotide sequence of the aspC gene". J. Biochem. 97 (4): 1259–62. PMID 3897210.
- Kondo K, Wakabayashi S, Yagi T, Kagamiyama H (1984). "The complete amino acid sequence of aspartate aminotransferase from Escherichia coli: sequence comparison with pig isoenzymes". Biochem. Biophys. Res. Commun. 122 (1): 62–67. doi:10.1016/0006-291X(84)90439-X. PMID 6378205.
- Inoue K, Kuramitsu S, Okamoto A, Hirotsu K, Higuchi T, Kagamiyama H (1991). "Site-directed mutagenesis of Escherichia coli aspartate aminotransferase: role of Tyr70 in the catalytic processes". Biochemistry 30 (31): 7796–7801. doi:10.1021/bi00245a019. PMID 1868057.
External links
Transferase: nitrogenous groups (EC 2.6) 2.6.1: Transaminases Aspartate transaminase (GOT1, GOT2) · Alanine transaminase · GABA transaminase · Tyrosine aminotransferase · Ornithine aminotransferase · Branched chain aminotransferase · Alanine-glyoxylate transaminase (AGXT)2.6.3: Oximinotransferases 2.6.99: Other Metabolism: Citric acid cycle enzymes Cycle Anaplerotic to acetyl-CoAPyruvate dehydrogenase complex (E1, E2, E3)
(regulated by Pyruvate dehydrogenase kinase and Pyruvate dehydrogenase phosphatase)to succinyl-CoAto oxaloacetateMitochondrial
electron transport chain/
oxidative phosphorylationPrimaryComplex I/NADH dehydrogenase · Complex II/Succinate dehydrogenase · Coenzyme Q · Complex III/Coenzyme Q - cytochrome c reductase · Cytochrome c · Complex IV/Cytochrome c oxidase
Coenzyme Q10 synthesis: COQ2 · COQ3 · COQ4 · COQ5 · COQ6 · COQ7 · COQ9 · COQ10A · COQ10B · PDSS1 · PDSS2OtherMetabolism: amino acid metabolism · synthesis and catabolism enzymes (essential in CAPS) K→acetyl-CoA (see below)G →alpha-ketoglutarate→TCAOther→succinyl-CoA→TCAG→fumarateasparagine→aspartate→Medical test: Serology, reference range: Clinical biochemistry blood tests (including BMP, CMP) (CPT 82000-84999) Fluid/electrolytes electrolytes (Na+/K+, Cl-/HCO3-) · renal function, BUN-to-creatinine ratio (BUN/Creatinine) · Ca
derived values: Plasma osmolality · Serum osmolal gapAcid-base Nutrition Iron tests: Transferrin saturation = Serum iron / Total iron-binding capacity; Ferritin · Transferrin · Transferrin receptorEndocrine ACTH stimulation test · Thyroid function tests (TSH)
Blood sugar: Glucose test · C-peptide · Fructosamine · Glycated hemoglobinMetabolic Cardiovascular Digestive Liver function tests: protein tests (Human serum albumin, Serum total protein) · ALP · transaminases (ALT, AST, AST/ALT ratio) · Bilirubin (Unconjugated, Conjugated)
Amylase · Lipase (Pancreatic lipase)Categories:- Liver function tests
- EC 2.6.1
Wikimedia Foundation. 2010.