- Cytochrome c
-
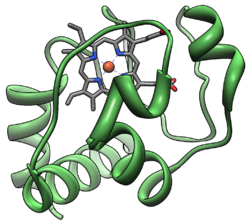
The Cytochrome complex, or cyt c is a small heme protein found loosely associated with the inner membrane of the mitochondrion. It belongs to the cytochrome c family of proteins. Cytochrome c is a highly soluble protein, unlike other cytochromes, with a solubility of about 100 g/L and is an essential component of the electron transport chain, where it carries one electron. It is capable of undergoing oxidation and reduction, but does not bind oxygen. It transfers electrons between Complexes III (Coenzyme Q - Cyt C reductase) and IV (Cyt C oxidase). In humans, cytochrome c is encoded by the CYCS gene.[1][2]
Contents
Function
Cytochrome c is a component of the electron transport chain in mitochondria. The heme group of cytochrome c accepts electrons from the b-c1 complex and transfers electrons to the cytochrome oxidase complex. Cytochrome c is also involved in initiation of apoptosis. Upon release of cytochrome c to the cytoplasm, the protein binds apoptotic protease activating factor.[1]
Cytochrome c can catalyze several reactions such as hydroxylation and aromatic oxidation, and shows peroxidase activity by oxidation of various electron donors such as 2,2-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS), 2-keto-4-thiomethyl butyric acid and 4-aminoantipyrine.
Species distribution
Cytochrome c is a highly conserved protein across the spectrum of species, found in plants, animals, and many unicellular organisms. This, along with its small size (molecular weight about 12,000 daltons), makes it useful in studies of cladistics. Its primary structure consists of a chain of about 100 amino acids. Many higher order organisms possess a chain of 104 amino acids.[3]
The cytochrome c molecule has been studied for the glimpse it gives into evolutionary biology. Both chickens and turkeys have identical sequence homology (amino acid for amino acid), whereas ducks possess molecules differing by one amino acid. Similarly, both humans and chimpanzees have the identical molecule, while rhesus monkeys share all but one of the amino acids:[4] the 66th amino acid is isoleucine in the former and threonine in the latter.[3] Pigs, cows and sheep also share identical cytochrome c molecules.[3]
Classes
In 1991 R. P. Ambler recognized four classes of cytochrome c:[5]
- Class I includes the lowspin soluble cytochrome c of mitochondria and bacteria. It has the heme-attachment site towards the N terminus of histidine and the sixth ligand provided by a methionine residue towards the C terminus.
- Class II includes the highspin cytochrome c'. It has the heme-attachment site closed to the N terminus of histidine.
- Class III comprises the low redox potential multiple heme cytochromes. The heme c groups are structurally and functionally nonequivalent and present different redox potentials in the range 0 to -400 mV.
- Class IV was originally created to hold the complex proteins that have other prosthetic groups as well as heme c.
Applications
Cytochrome c is suspected to be the functional complex in so called LLLT: Low-level laser therapy. In LLLT, red light and some near infra-red wavelengths penetrate tissue in order to increase cellular regeneration. Light of this wavelength appears capable of increasing activity of cytochrome c, thus increasing metabolic activity and freeing up more energy for the cells to repair the tissue.[6]
Interactions
Cytochrome c has been shown to interact with COX4I2.[7][8][9][10]
Role in apoptosis
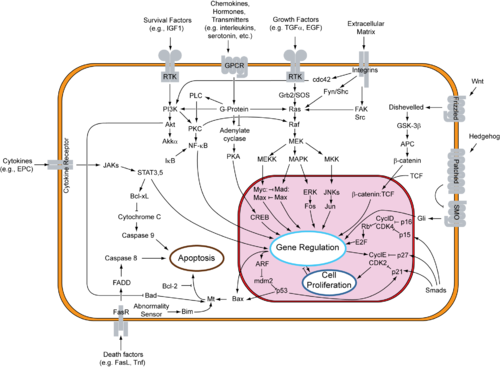
Cytochrome c is also an intermediate in apoptosis, a controlled form of cell death used to kill cells in the process of development or in response to infection or DNA damage.[11]
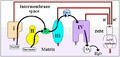
Cytochrome c is released by the mitochondria in response to pro-apoptotic stimuli. The sustained elevation in calcium levels precedes cyt c release from the mitochondria. The release of small amounts of cyt c leads to an interaction with the IP3 receptor (IP3R) on the endoplasmic reticulum (ER), causing ER calcium release. The overall increase in calcium triggers a massive release of cyt c, which then acts in the positive feedback loop to maintain ER calcium release through the IP3Rs. This explains how the ER calcium release can reach cytotoxic levels. This release of cytochrome c in turn activates caspase 9, a cysteine protease. Caspase 9 can then go on to activate caspase 3 and caspase 7, which are responsible for destroying the cell from within.
Cytochrome c binds to cardiolipin in the inner mitochondrial membrane, thus anchoring its presence and keeping it from releasing out of the mitochondria and initiating apoptosis. While the initial attraction between cardiolipin and cytochrome c is electrostatic due to the extreme positive charge on cytochrome c, the final interaction is hydrophobic, where a hydrophobic tail from cardiolipin inserts itself into the hydrophobic portion of cytochrome c.
See also
References
- ^ a b "Entrez Gene: cytochrome c". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=54205.
- ^ Tafani M, Karpinich NO, Hurster KA, Pastorino JG, Schneider T, Russo MA, Farber JL (March 2002). "Cytochrome c release upon Fas receptor activation depends on translocation of full-length bid and the induction of the mitochondrial permeability transition". J. Biol. Chem. 277 (12): 10073–82. doi:10.1074/jbc.M111350200. PMID 11790791.
- ^ a b c Amino acid sequences in cytochrome c proteins from different species, adapted from Strahler, Arthur; Science and Earth History, 1997. page 348.
- ^ Linda Stone; Paul F. Lurquin; Luigi Luca Cavalli-Sforza. Genes, Culture, and Human Evolution: A Synthesis, Blackwell Publishing, 2007, page 79. ISBN 1405150890
- ^ Ambler RP (May 1991). "Sequence variability in bacterial cytochromes c". Biochim. Biophys. Acta 1058 (1): 42–7. doi:10.1016/S0005-2728(05)80266-X. PMID 1646017.
- ^ Silveira PC, Streck EL, Pinho RA. (2005). "Cellular effects of low power laser therapy can be mediated by nitric oxide.". Lasers Surg Med. 36 (4): 307–14. doi:10.1002/lsm.20148. PMID 15739174.
- ^ Michel, B; Bosshard H R (Aug. 1984). "Spectroscopic analysis of the interaction between cytochrome c and cytochrome c oxidase". J. Biol. Chem. (UNITED STATES) 259 (16): 10085–91. ISSN 0021-9258. PMID 6088481.
- ^ Wiedemann, F R; Vielhaber S, Schröder R, Elger C E, Kunz W S (Mar. 2000). "Evaluation of methods for the determination of mitochondrial respiratory chain enzyme activities in human skeletal muscle samples". Anal. Biochem. (UNITED STATES) 279 (1): 55–60. doi:10.1006/abio.1999.4434. ISSN 0003-2697. PMID 10683230.
- ^ Sampson, V; Alleyne T (Dec. 2001). "Cytochrome c/cytochrome c oxidase interaction. Direct structural evidence for conformational changes during enzyme turnover". Eur. J. Biochem. (Germany) 268 (24): 6534–44. doi:10.1046/j.0014-2956.2001.02608.x. ISSN 0014-2956. PMID 11737208.
- ^ Lynch, S R; Sherman D, Copeland R A (Jan. 1992). "Cytochrome c binding affects the conformation of cytochrome a in cytochrome c oxidase". J. Biol. Chem. (UNITED STATES) 267 (1): 298–302. ISSN 0021-9258. PMID 1309738.
- ^ Liu X, Kim C, Yang J, Jemmerson R, Wang X (1996). "Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c". Cell 86 (1): 147–57. doi:10.1016/S0092-8674(00)80085-9. PMID 8689682.
Further reading
- Kumarswamy R and Chandna S (2009). "Putative partners in Bax mediated cytochrome-c release: ANT, CypD, VDAC or none of them?". Mitochondrion. 9 (1): 1–8. doi:10.1016/j.mito.2008.10.003. PMID 18992370.
- Skulachev VP (1998). "Cytochrome c in the apoptotic and antioxidant cascades.". FEBS Lett. 423 (3): 275–80. doi:10.1016/S0014-5793(98)00061-1. PMID 9515723.
- Mannella CA (1998). "Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications.". J. Struct. Biol. 121 (2): 207–18. doi:10.1006/jsbi.1997.3954. PMID 9615439.
- Ferri KF, Jacotot E, Blanco J et al. (2001). "Mitochondrial control of cell death induced by HIV-1-encoded proteins". Ann. N. Y. Acad. Sci. 926: 149–64. doi:10.1111/j.1749-6632.2000.tb05609.x. PMID 11193032.
- Britton RS, Leicester KL, Bacon BR (2002). "Iron toxicity and chelation therapy". Int. J. Hematol. 76 (3): 219–28. doi:10.1007/BF02982791. PMID 12416732.
- Haider N, Narula N, Narula J (2003). "Apoptosis in heart failure represents programmed cell survival, not death, of cardiomyocytes and likelihood of reverse remodeling". J. Card. Fail. 8 (6 Suppl): S512–7. doi:10.1054/jcaf.2002.130034. PMID 12555167.
- Castedo M, Perfettini JL, Andreau K et al. (2004). "Mitochondrial apoptosis induced by the HIV-1 envelope". Ann. N. Y. Acad. Sci. 1010: 19–28. doi:10.1196/annals.1299.004. PMID 15033690.
- Ng S, Smith MB, Smith HT, Millett F (1977). "Effect of modification of individual cytochrome c lysines on the reaction with cytochrome b5". Biochemistry 16 (23): 4975–8. doi:10.1021/bi00642a006. PMID 199233.
- Lynch SR, Sherman D, Copeland RA (1992). "Cytochrome c binding affects the conformation of cytochrome a in cytochrome c oxidase". J. Biol. Chem. 267 (1): 298–302. PMID 1309738.
- Garber EA, Margoliash E (1990). "Interaction of cytochrome c with cytochrome c oxidase: an understanding of the high- to low-affinity transition". Biochim. Biophys. Acta 1015 (2): 279–87. doi:10.1016/0005-2728(90)90032-Y. PMID 2153405.
- Bedetti CD (1985). "Immunocytochemical demonstration of cytochrome c oxidase with an immunoperoxidase method: a specific stain for mitochondria in formalin-fixed and paraffin-embedded human tissues". J. Histochem. Cytochem. 33 (5): 446–52. doi:10.1177/33.5.2580882. PMID 2580882.
- Tanaka Y, Ashikari T, Shibano Y et al. (1988). "Construction of a human cytochrome c gene and its functional expression in Saccharomyces cerevisiae". J. Biochem. 103 (6): 954–61. PMID 2844747.
- Evans MJ, Scarpulla RC (1989). "The human somatic cytochrome c gene: two classes of processed pseudogenes demarcate a period of rapid molecular evolution". Proc. Natl. Acad. Sci. U.S.A. 85 (24): 9625–9. doi:10.1073/pnas.85.24.9625. PMC 282819. PMID 2849112. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=282819.
- Passon PG, Hultquist DE (1972). "Soluble cytochrome b 5 reductase from human erythrocytes". Biochim. Biophys. Acta 275 (1): 62–73. doi:10.1016/0005-2728(72)90024-2. PMID 4403130.
- Dowe RJ, Vitello LB, Erman JE (1984). "Sedimentation equilibrium studies on the interaction between cytochrome c and cytochrome c peroxidase". Arch. Biochem. Biophys. 232 (2): 566–73. doi:10.1016/0003-9861(84)90574-5. PMID 6087732.
- Michel B, Bosshard HR (1984). "Spectroscopic analysis of the interaction between cytochrome c and cytochrome c oxidase". J. Biol. Chem. 259 (16): 10085–91. PMID 6088481.
- Broger C, Nałecz MJ, Azzi A (1980). "Interaction of cytochrome c with cytochrome bc1 complex of the mitochondrial respiratory chain". Biochim. Biophys. Acta 592 (3): 519–27. doi:10.1016/0005-2728(80)90096-1. PMID 6251869.
- Smith HT, Ahmed AJ, Millett F (1981). "Electrostatic interaction of cytochrome c with cytochrome c1 and cytochrome oxidase". J. Biol. Chem. 256 (10): 4984–90. PMID 6262312.
- Geren LM, Millett F (1981). "Fluorescence energy transfer studies of the interaction between adrenodoxin and cytochrome c". J. Biol. Chem. 256 (20): 10485–9. PMID 6270113.
- Favre B, Zolnierowicz S, Turowski P, Hemmings BA (1994). "The catalytic subunit of protein phosphatase 2A is carboxyl-methylated in vivo". J. Biol. Chem. 269 (23): 16311–7. PMID 8206937.
- Gao B, Eisenberg E, Greene L (1996). "Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate". J. Biol. Chem. 271 (28): 16792–7. doi:10.1074/jbc.271.28.16792. PMID 8663341.
Additional images
External links
- The Cytochrome c Protein
- Apoptosis & Caspase 3 - PMAP The Proteolysis Map-animation
- UMich Orientation of Proteins in Membranes families/superfamily-78 - Calculated orientations of cytochromes c in the lipid bilayer
- MeSH Cytochrome+c
PDB gallery Metabolism: Citric acid cycle enzymes Cycle Anaplerotic to acetyl-CoAPyruvate dehydrogenase complex (E1, E2, E3)
(regulated by Pyruvate dehydrogenase kinase and Pyruvate dehydrogenase phosphatase)to succinyl-CoAto oxaloacetateMitochondrial
electron transport chain/
oxidative phosphorylationPrimaryComplex I/NADH dehydrogenase · Complex II/Succinate dehydrogenase · Coenzyme Q · Complex III/Coenzyme Q - cytochrome c reductase · Cytochrome c · Complex IV/Cytochrome c oxidase
Coenzyme Q10 synthesis: COQ2 · COQ3 · COQ4 · COQ5 · COQ6 · COQ7 · COQ9 · COQ10A · COQ10B · PDSS1 · PDSS2OtherApoptosis signaling pathway Fas path IntracellularDeath-inducing signaling complex
DAXX · ASK1
Cytochrome c · Caspase 9 · Caspase 3
pro-apoptotic: BAX · BAK1/Bcl-2 homologous antagonist killer · Bcl-2-associated death promoter
anti-apoptotic: Bcl-2 · Bcl-xLTNF path IntracellularOther IntracellularCategories:- Human proteins
- Cellular respiration
- Cytochromes
- Programmed cell death
- Peripheral membrane proteins
- Moonlighting proteins
Wikimedia Foundation. 2010.