- Ornithine decarboxylase
-
ornithine decarboxylase Identifiers EC number 4.1.1.17 CAS number 9024-60-6 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles ornithine decarboxylase Identifiers Symbol ODC1 Entrez 4953 HUGO 8109 OMIM 165640 RefSeq NM_002539 UniProt P11926 Other data EC number 4.1.1.17 Locus Chr. 2 p25 The enzyme ornithine decarboxylase (ODC) catalyzes the decarboxylation of ornithine (a product of the urea cycle) to form putrescine. This reaction is the committed step in polyamine synthesis.[1] In humans, this protein has 461 amino acids and forms a homodimer.
Contents
Reaction mechanism
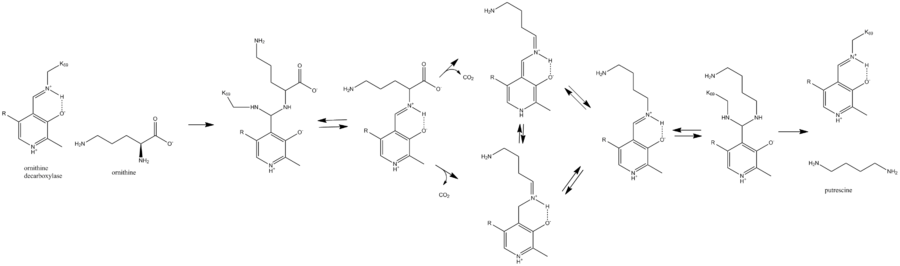
Lysine 69 on ornithine decarboxylase binds the cofactor pyridoxal phosphate to form a Schiff base. Ornithine displaces the lysine to form a Schiff base attached to ornithine, which decarboxylates to form a quinoid intermediate. This intermediate rearranges to form a Schiff base attached to putrescine, which is attacked by lysine to release putrescine product and reform PLP-bound ornithine decarboxylase.[2]
This is the first step and the rate-limiting step in humans for the production of polyamines, compounds required for cell division.
Structure
The active form of ornithine decarboyxlase is a homodimer. Each monomer contains a barrel domain, consisting of an alpha-beta barrel, and a sheet domain, composed of two beta-sheets. The domains are connected by loops. The monomers connect to each other via interactions between the barrel of one monomer and the sheet of the other. Binding between monomers is relatively weak, and ODC interconverts rapidly between monomeric and dimeric forms in the cell.[1]
The pyridoxal phosphate cofactor binds lysine 69 at the C-terminus end of the barrel domain. The active site is at the interface of the two domains, in a cavity formed by loops from both monomers.[1]
Function
The ornithine decarboxylation reaction catalyzed by ornithine decarboxylase is the first and committed step in the synthesis of polyamines, particularly putrescine, spermidine and spermine. Polyamines are important for stabilizing DNA structure, the DNA double strand-break repair pathway and as antioxidants. Therefore ornithine decarboxylase is an essential enzyme for cell growth, producing the polyamines necessary to stabilize newly synthesized DNA. Lack of ODC causes cell apoptosis in embryonic mice, induced by DNA damage.[4]
Proteasomal degradation
ODC is the most well-characterized cellular protein subject to ubiquitin-independent proteasomal degradation. Although most proteins must first be tagged with multiple ubiquitin molecules before they are bound and degraded by the proteasome, ODC degradation is instead mediated by several recognition sites on the protein and its accessory factor antizyme. The ODC degradation process is regulated in a negative feedback loop by its reaction products.[5]
Until a report by Sheaff et al. (2000),[6] which demonstrated that the cyclin-dependent kinase (Cdk) inhibitor p21Cip1 is also degraded by the proteasome in a ubiquitin-independent manner, ODC was the only clear example of ubiquitin-independent proteasomal degradation.[7]
Clinical significance
ODC is a transcriptional target of the oncogene Myc[8] and is upregulated in a wide variety of cancers. The polyamine products of the pathway initialized by ODC are associated with increased cell growth and reduced apoptosis.[9] Ultraviolet light,[10] asbestos[11] and androgens released by the prostate gland[12] are all known to induce increased ODC activity associated with cancer. Inhibitors of ODC such as eflornithine have been shown to effectively reduce cancers in animal models,[13] and drugs targeting ODC are being tested for potential clinical use. The mechanism by which ODC promotes carcinogenesis is complex and not entirely known. Along with their direct effect on DNA stability, polyamines also upregulate gap junction genes[14] and downregulate tight junction genes. Gap junction genes are involved in communication between carcinogenic cells and tight junction genes act as tumor suppressors.[9]
ODC gene expression is induced by a large number of biological stimuli including seizure activity in the brain.[15] Inactivation of ODC by difluoromethylornithine (eflornithine) is used to treat cancer and facial hair growth in postmenopausal females.
ODC is also an enzyme indispensable to parasites like trypanosoma, giardia, and plasmodium, a fact exploited by the drug eflornithine.[16]
References
- ^ a b c Kern AD, Oliveira MA, Coffino P, Hackert ML (May 1999). "Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases". Structure 7 (5): 567–81. doi:10.1016/S0969-2126(99)80073-2. PMID 10378276.
- ^ Brooks HB, Phillips MA (December 1997). "Characterization of the reaction mechanism for Trypanosoma brucei ornithine decarboxylase by multiwavelength stopped-flow spectroscopy". Biochemistry 36 (49): 15147–55. doi:10.1021/bi971652b. PMID 9398243.
- ^ PDB 1d7k; Almrud JJ, Oliveira MA, Kern AD, Grishin NV, Phillips MA, Hackert ML. (January 2000). "Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding". J. Mol. Biol. 295 (1): 7–16. doi:10.1006/jmbi.1999.3331. PMID 10623504. http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-45F4V3C-5P&_user=483702&_coverDate=01%2F07%2F2000&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_searchStrId=1468011589&_rerunOrigin=google&_acct=C000022720&_version=1&_urlVersion=0&_userid=483702&md5=1cea1cd3d0bf40355b774bdf274fe9e0&searchtype=a.; rendered via PyMOL.
- ^ Pendeville H, Carpino N, Marine JC, et al. (October 2001). "The ornithine decarboxylase gene is essential for cell survival during early murine development". Mol. Cell. Biol. 21 (19): 6549–58. doi:10.1128/MCB.21.19.6549-6558.2001. PMC 99801. PMID 11533243. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=99801.
- ^ Zhang M, Pickart CM, Coffino P (April 2003). "Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate". EMBO J. 22 (7): 1488–96. doi:10.1093/emboj/cdg158. PMC 152902. PMID 12660156. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=152902.
- ^ Sheaff RJ, Singer JD, Swanger J, Smitherman M, Roberts JM, Clurman BE (February 2000). "Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination". Mol. Cell 5 (2): 403–10. doi:10.1016/S1097-2765(00)80435-9. PMID 10882081.
- ^ Verma R, Deshaies RJ (May 2000). "A proteasome howdunit: the case of the missing signal". Cell 101 (4): 341–4. doi:10.1016/S0092-8674(00)80843-0. PMID 10830160.
- ^ Bello-Fernandez C, Packham G, Cleveland JL (August 1993). "The ornithine decarboxylase gene is a transcriptional target of c-Myc". Proc. Natl. Acad. Sci. U.S.A. 90 (16): 7804–8. doi:10.1073/pnas.90.16.7804. PMC 47231. PMID 8356088. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=47231.
- ^ a b Gerner EW, Meyskens FL (October 2004). "Polyamines and cancer: old molecules, new understanding". Nat. Rev. Cancer 4 (10): 781–92. doi:10.1038/nrc1454. PMID 15510159.
- ^ Ahmad N, Gilliam AC, Katiyar SK, O'Brien TG, Mukhtar H (September 2001). "A definitive role of ornithine decarboxylase in photocarcinogenesis". Am. J. Pathol. 159 (3): 885–92. doi:10.1016/S0002-9440(10)61764-6. PMC 1850478. PMID 11549581. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1850478.
- ^ Marsh JP, Mossman BT (January 1991). "Role of asbestos and active oxygen species in activation and expression of ornithine decarboxylase in hamster tracheal epithelial cells". Cancer Res. 51 (1): 167–73. PMID 1846307.
- ^ Crozat A, Palvimo JJ, Julkunen M, Jänne OA (March 1992). "Comparison of androgen regulation of ornithine decarboxylase and S-adenosylmethionine decarboxylase gene expression in rodent kidney and accessory sex organs". Endocrinology 130 (3): 1131–44. doi:10.1210/en.130.3.1131. PMID 1537280.
- ^ Meyskens FL, Gerner EW (May 1999). "Development of difluoromethylornithine (DFMO) as a chemoprevention agent". Clin. Cancer Res. 5 (5): 945–51. PMID 10353725.
- ^ Shore L, McLean P, Gilmour SK, Hodgins MB, Finbow ME (July 2001). "Polyamines regulate gap junction communication in connexin 43-expressing cells". Biochem. J. 357 (Pt 2): 489–95. doi:10.1042/0264-6021:3570489. PMC 1221976. PMID 11439099. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1221976.
- ^ Herberg LJ, Rose IC, de Belleroche JS, Mintz M (1992). "Ornithine decarboxylase induction and polyamine synthesis in the kindling of seizures: the effect of alpha-difluoromethylornithine". Epilepsy Res. 11 (1): 3–7. doi:10.1016/0920-1211(92)90015-L. PMID 1563337.
- ^ Heby O, Persson L, Rentala M (August 2007). "Targeting the polyamine biosynthetic enzymes: a promising approach to therapy of African sleeping sickness, Chagas' disease, and leishmaniasis". Amino Acids 33 (2): 359–66. doi:10.1007/s00726-007-0537-9. PMID 17610127.
External links
Carbon-carbon lyases (EC 4.1) 4.1.1: Carboxy-lyases Pyruvate decarboxylase · Oxaloacetate decarboxylase · Acetoacetate decarboxylase · Malonyl-CoA decarboxylase · Glutamate decarboxylase · Ornithine decarboxylase · Lysine decarboxylase · Phosphoribosylaminoimidazole carboxylase · Histidine decarboxylase · Uridine monophosphate synthetase/Orotidine 5'-phosphate decarboxylase · Aromatic L-amino acid decarboxylase · Phosphoenolpyruvate carboxylase · Pyrophosphomevalonate decarboxylase · Uroporphyrinogen III decarboxylase · RuBisCO · Phosphoenolpyruvate carboxykinase · Adenosylmethionine decarboxylase4.1.2: Aldehyde-lyases 4.1.3: Oxo-acid-lyases 4.1.99: Other B enzm: 1.1/2/3/4/5/6/7/8/10/11/13/14/15-18, 2.1/2/3/4/5/6/7/8, 2.7.10, 2.7.11-12, 3.1/2/3/4/5/6/7, 3.1.3.48, 3.4.21/22/23/24, 4.1/2/3/4/5/6, 5.1/2/3/4/99, 6.1-3/4/5-6 Metabolism: amino acid metabolism · synthesis and catabolism enzymes (essential in CAPS) K→acetyl-CoA (see below)G →alpha-ketoglutarate→TCAOther→succinyl-CoA→TCAG→fumarateasparagine→aspartate→Categories:- Genes on chromosome 2
- EC 4.1.1
- Enzymes
Wikimedia Foundation. 2010.