- Pyridoxal phosphate
-
Pyridoxal phosphate 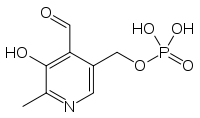
 [(4-formyl-5-hydroxy-6-methylpyridin-3-yl)methoxy]phosphonic acidOther namesPyridoxal 5-phosphate, PAL-P, PLP, Vitamin B6 phosphate[1]
[(4-formyl-5-hydroxy-6-methylpyridin-3-yl)methoxy]phosphonic acidOther namesPyridoxal 5-phosphate, PAL-P, PLP, Vitamin B6 phosphate[1]Identifiers CAS number 54-47-7 
PubChem 1051 MeSH Pyridoxal+Phosphate ChEMBL CHEMBL82202 
Jmol-3D images Image 1 - CC1=NC=C(C(=C1O)
C=O)COP(=O)(O)O
Properties Molecular formula C8H10NO6P Molar mass 247.142 g/mol Density 1.638±0.06 g/cm3[2] Melting point 139-142°C[3]
Acidity (pKa) 1.56[4] Hazards Flash point 296.0±32.9 °C[5]  phosphate (verify) (what is:
phosphate (verify) (what is:  /
/ ?)
?)
Except where noted otherwise, data are given for materials in their standard state (at 25 °C, 100 kPa)Infobox references Pyridoxal-phosphate (PLP, pyridoxal-5'-phosphate, P5P) is a prosthetic group of some enzymes. It is the active form of vitamin B6, which comprises three natural organic compounds, pyridoxal, pyridoxamine and pyridoxine.
Contents
Role as a coenzyme
PLP acts as a coenzyme in all transamination reactions, and in some decarboxylation and deamination reactions of amino acids. The aldehyde group of PLP forms a Schiff-base linkage (internal aldimine) with the ε-amino group of a specific lysine group of the aminotransferase enzyme. The α-amino group of the amino acid substrate displaces the ε-amino group of the active-site lysine residue. The resulting external aldimine becomes deprotonated to become a quinoid intermediate, which in turn accepts a proton at a different position to become a ketimine. The resulting ketimine is hydrolysed so that the amino group remains on the complex.[6]
In addition, PLP is used by aminotransferases (or transaminases) that act upon unusual sugars such as perosamine and desosamine.[7] In these reactions, the PLP reacts with glutamate, which transfers its alpha-amino group to PLP to make pyridoxamine phosphate (PMP). PMP then transfers its nitrogen to the sugar, making an amino sugar.
PLP is also involved in various beta-elimination reactions such as the reactions carried out by serine dehydratase and GDP-4-keto-6-deoxymannose-3-dehydratase (ColD).[7]
It is also active in the condensation reaction in heme synthesis.
PLP plays a role in the conversion of dopa into dopamine, allows the conversion of the excitatory neurotransmitter glutamate to the inhibitory neurotransmitter GABA, and allows SAM to be decarboxylated to form propylamine, which is a precursor to polyamines.
Non-classical examples of PLP
PLP is also found on glycogen phosphorylase in the liver, where it is used to break down glycogen in glycogenolysis when glucagon or epinephrine signals it to do so. However, this enzyme does not exploit the reactive aldehyde group, but instead utilizes the phosphate group on PLP to perform its reaction.
Although the vast majority of PLP-dependent enzymes form an internal aldimine with PLP via an active site lysine residue; some PLP-dependent enzymes do not have this lysine residue, but instead have an active site histidine. In such a case, the histidine cannot form the internal aldimine, and, therefore, the cofactor never becomes covalently tethered to the enzyme. GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) is an example of such an enzyme.[8]
Catalytic Mechanism
The pyridoxal-5′-phosphate-dependent enzymes (PLP enzymes) catalyze myriad biochemical reactions. Although the scope of PLP-catalyzed reactions initially appears to be immensely diverse, there is a simple unifying principle: In the resting state, the cofactor (PLP) is covalently bonded to the amino group of an active site lysine, forming an internal aldimine. Once the amino substrate interacts with the active site, a new Schiff base is generated, commonly referred to as the external aldimine. Only after this step, the mechanistic pathway for each PLP-catalyzed reaction diverges. Density functional methods have been applied to investigate the transimination reaction, and the results have shown that the reaction involves three sequential steps: (i) formation of a tetrahedral intermediate with the active site lysine and the amino substrate bonded to the PLP cofactor; (ii) nondirect proton transfer between the amino substrate and the lysine residue; and (iii) formation of the external aldimine after the dissociation of the lysine residue. The overall reaction is exothermic (−12.0 kcal/mol), and the rate-limiting step is the second one with 12.6 kcal/mol for the activation energy[9]
Biological Synthesis
It is synthesized from pyridoxal by the enzyme pyridoxal kinase, requiring one ATP. It is metabolized in the liver.
See also
- Aromatic-L-amino-acid decarboxylase
- Ornithine decarboxylase
References
- ^ Anonymous . Substance Detail. https://scifinder-cas-org.proxy.library.nd.edu:9443/scifinder/view/scifinder/scifinderExplore.jsf (accessed 12 Nov, 2011).
- ^ Calculated using Advanced Chemistry Development (ACD/Labs) Software V11.02 (© 1994-2011 ACD/Labs)
- ^ Kozlov E.I., L. M. S. Stability of water-soluble vitamins and coenzymes. Hydrolysis of pyridoxal-5-phosphate in acidic, neutral, and weakly alkaline solutions. Pharmaceutical Chemistry Journal 1978, 11, 1543.
- ^ Calculated using Advanced Chemistry Development (ACD/Labs) Software V11.02 (© 1994-2011 ACD/Labs)
- ^ Calculated using Advanced Chemistry Development (ACD/Labs) Software V11.02 (© 1994-2011 ACD/Labs)
- ^ Toney, M. D. "Reaction specificity in pyridoxal enzymes." Archives of biochemistry and biophysics (2005) 433: 279-287.
- ^ a b Samuel, G. and Reeves, P. "Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precusor synthesis and O-antigen assembly." Carbohydrate research (2003) 338:2503-2519.
- ^ Cook P. D., Thoden J.B. and Holden H. M. "The structure of GDP-4-keto-6-deoxymannose-3-dehydratase: a unique coenzyme B6-dependent enzyme." Protein Science (2006) 15:2093-2106.
- ^ N. M. F. S. A. Cerqueira, P. A. Fernandes, M. J. Ramos (2011). "Computational Mechanistic Studies Addressed to the Transimination Reaction Present in All Pyridoxal 5′-Phosphate-Requiring Enzymes". Journal of Chemical Theory and Computation 7 (5): 1356–1368. doi:10.1021/ct1002219.
External links
Vitamins (A11) Fat soluble D2 (Ergosterol, Ergocalciferol#) · D3 (7-Dehydrocholesterol, Previtamin D3, Cholecalciferol, 25-hydroxycholecalciferol, Calcitriol (1,25-dihydroxycholecalciferol), Calcitroic acid) · D4 (Dihydroergocalciferol) · D5 · D analogues (Dihydrotachysterol, Calcipotriol, Tacalcitol, Paricalcitol)Water soluble B1 (Thiamine#) · B2 (Riboflavin#) · B3 (Niacin, Nicotinamide#) · B5 (Pantothenic acid, Dexpanthenol, Pantethine) · B6 (Pyridoxine#, Pyridoxal phosphate, Pyridoxamine) · B7 (Biotin) · B9 (Folic acid, Dihydrofolic acid, Folinic acid) · B12 (Cyanocobalamin, Hydroxocobalamin, Methylcobalamin, Cobamamide) · CholineCombinations M: NUT
cof, enz, met
noco, nuvi, sysi/epon, met
drug(A8/11/12)
Enzyme cofactors Active forms TPP / ThDP (B1) · FMN, FAD (B2) · NAD+, NADH, NADP+, NADPH (B3) · Coenzyme A (B5) · PLP / P5P (B6) · Biotin (B7) · THFA / H4FA, DHFA / H2FA, MTHF (B9) · AdoCbl, MeCbl (B12) · Ascorbic Acid (C) · Phylloquinone (K1), Menaquinone (K2) · Coenzyme F420ATP · CTP · SAMe · PAPS · GSH · Coenzyme B · Cofactor F430 · Coenzyme M · Coenzyme Q · Heme / Haem (A, B, C, O) · Lipoic Acid · Methanofuran · Molybdopterin/Molybdenum cofactor · PQQ · THB / BH4 · THMPT / H4MPTBase forms vitamins: see vitaminsM: NUT
cof, enz, met
noco, nuvi, sysi/epon, met
drug(A8/11/12)
Categories: - CC1=NC=C(C(=C1O)
Wikimedia Foundation. 2010.
