- Lysozyme
-
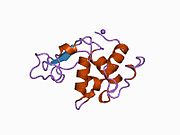
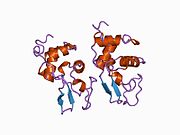
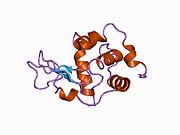
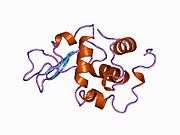
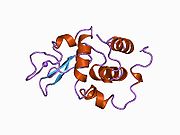
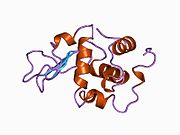
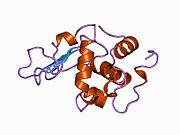
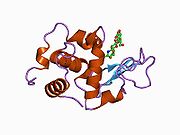
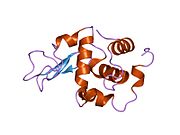
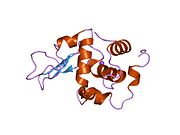
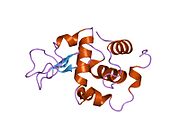
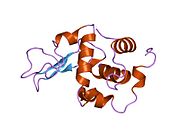
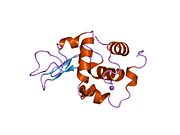
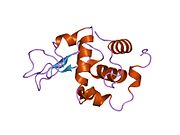
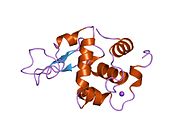
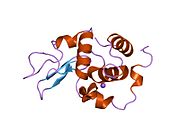
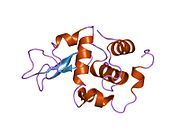
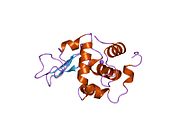
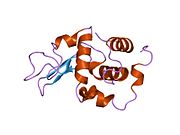
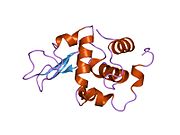
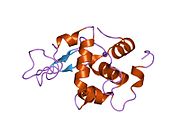
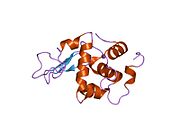
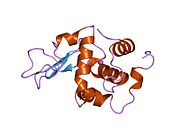
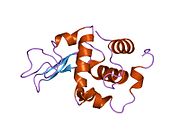
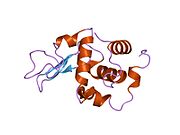
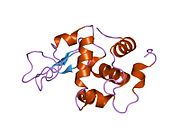
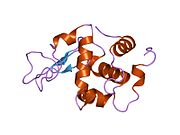
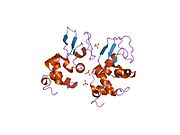
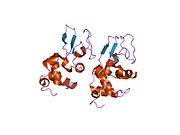
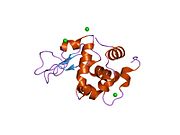
Lysozyme Lysozyme single crystal Identifiers EC number 3.2.1.17 CAS number 9001-63-2 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles Lysozyme, also known as muramidase or N-acetylmuramide glycanhydrolase, are glycoside hydrolases, enzymes (EC 3.2.1.17) that damage bacterial cell walls by catalyzing hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in a peptidoglycan and between N-acetyl-D-glucosamine residues in chitodextrins. Lysozyme is abundant in a number of secretions, such as tears, saliva, human milk, and mucus. It is also present in cytoplasmic granules of the polymorphonuclear neutrophils (PMN). Large amounts of lysozyme can be found in egg white. C-type lysozymes are closely related to alpha-lactalbumin in sequence and structure, making them part of the same family.
In humans, the lysozyme enzyme is encoded by the LYZ gene.[1][2]
Contents
Function
The enzyme functions by attacking peptidoglycans (found in the cell walls of bacteria, especially Gram-positive bacteria) and hydrolyzing the glycosidic bond that connects N-acetylmuramic acid with the fourth carbon atom of N-acetylglucosamine. It does this by binding to the peptidoglycan molecule in the binding site within the prominent cleft between its two domains. This causes the substrate molecule to adopt a strained conformation similar to that of the transition state[citation needed]. According to Phillips-Mechanism, the lysozyme binds to a hexasaccharide. The lysozyme then distorts the fourth sugar in hexasaccharide (the D ring) into a half-chair conformation. In this stressed state, the glycosidic bond is easily broken.
The amino acid side-chains glutamic acid 35 (Glu35) and aspartate 52 (Asp52) have been found to be critical to the activity of this enzyme. Glu35 acts as a proton donor to the glycosidic bond, cleaving the C-O bond in the substrate, whereas Asp52 acts as a nucleophile to generate a glycosyl enzyme intermediate. The glycosyl enzyme intermediate then reacts with a water molecule, to give the product of hydrolysis and leaving the enzyme unchanged.[citation needed]
Role in disease
Lysozyme is part of the innate immune system. Reduced lysozyme levels have been associated with bronchopulmonary dysplasia in newborns.[3] Children fed infant formula lacking lysozyme in their diet have three times the rate of diarrheal disease.[4] Since lysozyme is a natural form of protection from gram-positive pathogens like Bacillus and Streptococcus,[5] a deficiency due to infant formula feeding can lead to increased incidence of disease. Whereas the skin is a protective barrier due to its dryness and acidity, the conjunctiva (membrane covering the eye) is, instead, protected by secreted enzymes, mainly lysozyme and defensin. However, when these protective barriers fail, conjunctivitis results.
In certain cancers (especially myelomonocytic leukemia) excessive production of lysozyme by cancer cells can lead to toxic levels of lysozyme in the blood. High lysozyme blood levels can lead to kidney failure and low blood potassium, conditions that may improve or resolve with treatment of the primary malignancy
History
The antibacterial property of hen egg white, due to the lysozyme it contains, was first observed by Laschtschenko in 1909,[6] although it was not until 1922 that the name 'lysozyme' was coined, by Alexander Fleming (1881–1955), the discoverer of penicillin.[7] Fleming first observed the antibacterial action of lysozyme when he treated bacterial cultures with nasal mucus from a patient suffering from a head cold.[7]
The three-dimensional structure of hen egg white lysozyme was described by David Chilton Phillips (1924–1999) in 1965, when he obtained the first 2-Ångström (200 pm) resolution model via X-ray crystallography.[8][9] The structure was publicly presented at a Royal Institution lecture in 1965.[10] Lysozyme was the second protein structure and the first enzyme structure to be solved via X-ray diffraction methods, and the first enzyme to be fully sequenced that contains all twenty common amino acids.[11] As a result of Phillips' elucidation of the structure of lysozyme, it was also the first enzyme to have a detailed, specific mechanism suggested for its method of catalytic action.[12] This work led Phillips to provide an explanation for how enzymes speed up a chemical reaction in terms of its physical structures. The original mechanism proposed by Phillips was more recently revised.[13]
Howard Florey (1898–1968) and Ernst B. Chain (1906–1979) also investigated lysozymes. Although they never made much progress in this field, they, along with Fleming, developed penicillin.
See also
References
- ^ Yoshimura K, Toibana A, Nakahama K (January 1988). "Human lysozyme: sequencing of a cDNA, and expression and secretion by Saccharomyces cerevisiae". Biochem. Biophys. Res. Commun. 150 (2): 794–801. doi:10.1016/0006-291X(88)90461-5. PMID 2829884.
- ^ Peters CW, Kruse U, Pollwein R, Grzeschik KH, Sippel AE (July 1989). "The human lysozyme gene. Sequence organization and chromosomal localization". Eur. J. Biochem. 182 (3): 507–16. doi:10.1111/j.1432-1033.1989.tb14857.x. PMID 2546758.
- ^ Revenis ME, Kaliner MA (August 1992). "Lactoferrin and lysozyme deficiency in airway secretions: association with the development of bronchopulmonary dysplasia". J. Pediatr. 121 (2): 262–70. doi:10.1016/S0022-3476(05)81201-6. PMID 1640295.
- ^ Lönnerdal B (June 2003). "Nutritional and physiologic significance of human milk proteins". Am. J. Clin. Nutr. 77 (6): 1537S–1543S. PMID 12812151.
- ^ Microbiology: A human perspective. Nester, Anderson, Roberts, Nester. 5th Ed. 2007
- ^ Laschtschenko P (1909). "Über die keimtötende und entwicklungshemmende Wirkung Hühnereiweiß" (in German). Z. Hyg. InfektKrankh. 64: 419–427. doi:10.1007/BF02216170.
- ^ a b Fleming A (1 May 1922). "On a remarkable bacteriolytic element found in tissues and secretions". Proc Roy Soc Ser B 93 (653): 306–317. doi:10.1098/rspb.1922.0023. http://www.jstor.org/pss/80959.
- ^ Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. (1965). "Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution". Nature 206 (986): 757–61. doi:10.1038/206757a0. PMID 5891407.
- ^ Johnson LN, Phillips DC. (1965). "Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Angstrom resolution". Nature 206 (986): 761–3. doi:10.1038/206761a0. PMID 5840126.
- ^ Johnson, LN (1998). "The early history of lysozme". Nat Struct Mol Biol 5 (11): 942–944. doi:10.1038/2917. PMID 9808036.
- ^ Canfield, RE (1963). "The Amino Acid Sequence of Egg White Lysozyme". J Biol Chem 238 (8): 2698–2707. PMID 14063294. http://www.jbc.org/content/238/8/2698.short.
- ^ Proc R Soc Lond B Bio 167 (1009): 389–401. 1967.
- ^ Vocadlo DJ, Davies GJ, Laine R, Withers SG. (2001). "Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate". Nature 412 (6849): 835–8. doi:10.1038/35090602. PMID 11518970.
External links
- MeSH Muramidase
- "Lysozyme: enzyme, sequence, crystallization, structure". lysozyme.co.uk. http://lysozyme.co.uk/. Retrieved 2009-01-04.
- Proteopedia.org HEW Lysozyme
PDB gallery 133l: ROLE OF ARG 115 IN THE CATALYTIC ACTION OF HUMAN LYSOZYME. X-RAY STRUCTURE OF HIS 115 AND GLU 115 MUTANTS134l: ROLE OF ARG 115 IN THE CATALYTIC ACTION OF HUMAN LYSOZYME. X-RAY STRUCTURE OF HIS 115 AND GLU 115 MUTANTS1b5u: CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT1b5v: CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS1b5w: CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS1b5x: Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and x-ray analysis of six ser->ala mutants1b5y: CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS1b5z: CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS1b7l: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7m: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7n: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7o: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7p: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7q: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7r: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1b7s: VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES1bb3: HUMAN LYSOZYME MUTANT A96L1bb4: HUMAN LYSOZYME DOUBLE MUTANT A96L, W109H1bb5: HUMAN LYSOZYME MUTANT A96L COMPLEXED WITH CHITOTRIOSE1c43: MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES1c45: MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES1c46: MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES1c7p: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME WITH FOUR EXTRA RESIDUES (EAEA) AT THE N-TERMINAL1cj6: T11A MUTANT HUMAN LYSOZYME1cj7: T11V MUTANT HUMAN LYSOZYME1cj8: T40A MUTANT HUMAN LYSOZYME1cj9: T40V MUTANT HUMAN LYSOZYME1ckc: T43A MUTANT HUMAN LYSOZYME1ckd: T43V MUTANT HUMAN LYSOZYME1ckf: T52A MUTANT HUMAN LYSOZYME1ckg: T52V MUTANT HUMAN LYSOZYME1ckh: T70V MUTANT HUMAN LYSOZYME1d6p: HUMAN LYSOZYME L63 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL N,N'-DIACETYLCHITOBIOSE1d6q: HUMAN LYSOZYME E102 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL GLYCOSIDE OF N-ACETYLLACTOSAMINE1di3: ROLE OF AMINO ACID RESIDUES AT TURNS IN THE CONFORMATIONAL STABILITY AND FOLDING OF HUMAN LYSOZYME1di4: ROLE OF AMINO ACID RESIDUES AT TURNS IN THE CONFORMATIONAL STABILITY AND FOLDING OF HUMAN LYSOZYME1di5: ROLE OF AMINO ACID RESIDUES AT TURNS IN THE CONFORMATIONAL STABILITY AND FOLDING OF HUMAN LYSOZYME1eq4: CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME1eq5: CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME1eqe: CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME1gay: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gaz: Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions1gb0: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb2: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb3: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb5: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb6: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb7: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb8: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gb9: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gbo: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gbw: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gbx: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gby: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gbz: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gdw: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1gdx: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1ge0: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1ge1: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1ge2: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1ge3: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1ge4: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT LEFT-HANDED HELICAL POSITIONS1gev: BURIED POLAR MUTANT HUMAN LYSOZYME1gez: BURIED POLAR MUTANT HUMAN LYSOZYME1gf0: BURIED POLAR MUTANT HUMAN LYSOZYME1gf3: BURIED POLAR MUTANT HUMAN LYSOZYME1gf4: BURIED POLAR MUTANT HUMAN LYSOZYME1gf5: BURIED POLAR MUTANT HUMAN LYSOZYME1gf6: BURIED POLAR MUTANT HUMAN LYSOZYME1gf7: BURIED POLAR MUTANT HUMAN LYSOZYME1gf8: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gf9: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfa: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfe: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfg: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfh: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfj: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfk: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfr: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gft: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfu: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1gfv: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1hnl: CRYSTAL STRUCTURE OF A GLUTATHIONYLATED HUMAN LYSOZYME: A FOLDING INTERMEDIATE MIMIC IN THE FORMATION OF A DISULFIDE BOND1i1z: MUTANT HUMAN LYSOZYME (Q86D)1i20: MUTANT HUMAN LYSOZYME (A92D)1i22: MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D)1inu: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS1ioc: CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME, EAEA-I56T1ip1: G37A HUMAN LYSOZYME1ip2: G48A HUMAN LYSOZYME1ip3: G68A HUMAN LYSOZYME1ip4: G72A HUMAN LYSOZYME1ip5: G105A HUMAN LYSOZYME1ip6: G127A HUMAN LYSOZYME1ip7: G129A HUMAN LYSOZYME1iwt: Crystal Structure Analysis of Human lysozyme at 113K.1iwu: Crystal Structure Analysis of Human lysozyme at 127K.1iwv: Crystal Structure Analysis of Human lysozyme at 147K.1iww: Crystal Structure Analysis of Human lysozyme at 152K.1iwx: Crystal Structure Analysis of Human lysozyme at 161K.The maximum number of images (100) is exceeded ! Hydrolase: sugar hydrolases (EC 3.2) 3.2.1: Glycoside hydrolases Cellulase · Alpha-glucosidase (Acid, Neutral AB, Neutral C) · Beta-glucosidase (cytosolic) · Debranching enzymeOtherAmylase (Alpha-Amylase) · Chitinase · Lysozyme · Neuraminidase (NEU1, NEU2, NEU3, NEU4, Bacterial neuraminidase, Viral neuraminidase) · Galactosidases (Alpha, Beta) · alpha-Mannosidase · Glucuronidase · Hyaluronidase · Pullulanase · Glucosylceramidase (lysosomal, non-lysosomal) · Galactosylceramidase · Alpha-N-acetylgalactosaminidase (NAGA) · Alpha-N-acetylglucosaminidase · Fucosidase · Hexosaminidase (HEXA, HEXB) · Iduronidase · Maltase-glucoamylase · Heparanase (HPSE2)3.2.2: Hydrolysing
N-Glycosyl compoundsB enzm: 1.1/2/3/4/5/6/7/8/10/11/13/14/15-18, 2.1/2/3/4/5/6/7/8, 2.7.10, 2.7.11-12, 3.1/2/3/4/5/6/7, 3.1.3.48, 3.4.21/22/23/24, 4.1/2/3/4/5/6, 5.1/2/3/4/99, 6.1-3/4/5-6 Antimicrobial peptides: Granulocyte granule contents Azurophilic granules (1°) Myeloperoxidase · Defensins · neutral serine proteases (Proteinase 3) · Lysozyme · Bactericidal/permeability increasing protein · CollagenaseSpecific granules (2°) Antibacterials: cell envelope antibiotics (J01C-J01D) Intracellular inhibit peptidoglycan subunit synthesis and transport: NAM synthesis inhibition (Fosfomycin) • DADAL/AR inhibitors (Cycloserine) • bactoprenol inhibitors (Bacitracin)Glycopeptide inhibit PG chain elongation: Vancomycin# (Oritavancin, Telavancin) • Teicoplanin (Dalbavancin) • Ramoplaninβ-lactams/
(inhibit PBP
cross-links)aminopenicillins: Amoxicillin# • Ampicillin# (Pivampicillin, Hetacillin, Bacampicillin, Metampicillin, Talampicillin) • Epicillin
carboxypenicillins: Carbenicillin (Carindacillin) • Ticarcillin • Temocillin
ureidopenicillins: Azlocillin • Piperacillin • Mezlocillin
other: Mecillinam (Pivmecillinam) • SulbenicillinBenzylpenicillin (G)#: Clometocillin • Benzathine benzylpenicillin# • Procaine benzylpenicillin# • Azidocillin • Penamecillin
Phenoxymethylpenicillin (V)#: Propicillin • Benzathine phenoxymethylpenicillin • PheneticillinPenemsCefazolin# • Cefacetrile • Cefadroxil • Cefalexin • Cefaloglycin • Cefalonium • Cefaloridine • Cefalotin • Cefapirin • Cefatrizine • Cefazedone • Cefazaflur • Cefradine • Cefroxadine • CeftezoleCefaclor • Cefamandole • Cefminox • Cefonicid • Ceforanide • Cefotiam • Cefprozil • Cefbuperazone • Cefuroxime • Cefuzonam • cephamycin (Cefoxitin, Cefotetan, Cefmetazole) • carbacephem (Loracarbef)Cefixime# • Ceftriaxone# • antipseudomonal (Ceftazidime# • Cefoperazone) • Cefcapene • Cefdaloxime • Cefdinir • Cefditoren • Cefetamet • Cefmenoxime • Cefodizime • Cefotaxime • Cefpimizole • Cefpiramide • Cefpodoxime • Cefsulodin • Cefteram • Ceftibuten • Ceftiolene • Ceftizoxime • oxacephem (Flomoxef, Latamoxef ‡)4th (antips-)CombinationsOther polymyxins/detergent (Colistin, Polymyxin B) • depolarizing (Daptomycin) • hydrolyze NAM-NAG (Lysozyme) • Gramicidin#WHO-EM. ‡Withdrawn from market. Clinical trials: †Phase III. §Never to phase III Categories:- Human proteins
- EC 3.2.1
Wikimedia Foundation. 2010.