- Oxoguanine glycosylase
-
8-Oxoguanine glycosylase also known as OGG1 is a DNA glycosylase enzyme that, in humans, is encoded by the OGG1 gene. It is involved in base excision repair.
Contents
Function
OGG1 is the primary enzyme responsible for the excision of 7,8-dihydro-8-oxoguanine (8-oxoG), a mutagenic base byproduct that occurs as a result of exposure to reactive oxygen species (ROS). OGG1 is a bifunctional glycosylase, as it is able to both cleave the glycosidic bond of the mutagenic lesion and cause a strand break in the DNA backbone. Alternative splicing of the C-terminal region of this gene classifies splice variants into two major groups, type 1 and type 2, depending on the last exon of the sequence. Type 1 alternative splice variants end with exon 7 and type 2 end with exon 8. All variants have the N-terminal region in common. Many alternative splice variants for this gene have been described, but the full-length nature for every variant has not been determined. The N-terminus of this gene contains a mitochondrial targeting signal, essential for mitochondrial localization.[1]
Despite the presumed importance of this enzyme, mice lacking Ogg1 have been generated and found to have a normal lifespan,[2] and despite some early reports, do not show increased mutagenesis or cancer incidence.[citation needed][dubious ] There is some controversy as to whether deletion of Ogg1 actually leads to increased 8-oxo-dG levels: the HPLC-EC assay suggests up to 6 fold higher levels of 8-oxo-dG in nuclear DNA and 20-fold higher in mitochondrial DNA whereas the fappy-glycosylase assay indicates no change.[citation needed]
Interactions
Oxoguanine glycosylase has been shown to interact with XRCC1[3] and PKC alpha.[4]
References
- ^ "Entrez Gene: OGG1 8-oxoguanine DNA glycosylase". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4968.
- ^ Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE (November 1999). "Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage". Proc. Natl. Acad. Sci. U.S.A. 96 (23): 13300–5. doi:10.1073/pnas.96.23.13300. PMC 23942. PMID 10557315. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=23942.
- ^ Marsin, Stéphanie; Vidal Antonio E, Sossou Marguerite, Ménissier-de Murcia Josiane, Le Page Florence, Boiteux Serge, de Murcia Gilbert, Radicella J Pablo (Nov. 2003). "Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1". J. Biol. Chem. (United States) 278 (45): 44068–74. doi:10.1074/jbc.M306160200. ISSN 0021-9258. PMID 12933815.
- ^ Dantzer, Françoise; Luna Luisa, Bjørås Magnar, Seeberg Erling (Jun. 2002). "Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo". Nucleic Acids Res. (England) 30 (11): 2349–57. doi:10.1093/nar/30.11.2349. PMC 117190. PMID 12034821. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=117190.
Further reading
- Boiteux S, Radicella JP (2000). "The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis.". Arch. Biochem. Biophys. 377 (1): 1–8. doi:10.1006/abbi.2000.1773. PMID 10775435.
- Park J, Chen L, Tockman MS et al. (2004). "The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA repair enzyme and its association with lung cancer risk". Pharmacogenetics 14 (2): 103–9. doi:10.1097/00008571-200402000-00004. PMID 15077011.
- Hung RJ, Hall J, Brennan P, Boffetta P (2006). "Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review". Am. J. Epidemiol. 162 (10): 925–42. doi:10.1093/aje/kwi318. PMID 16221808.
- Mirbahai L, Kershaw RM, Green RM, Hayden RE, Meldrum RA, Hodges NJ. (2010). "Use of a molecular beacon to track the activity of base excision repair protein OGG1 in live cells". DNA Repair 9 (2): 144–152. doi:10.1016/j.dnarep.2009.11.009. PMID 20042377.
External links
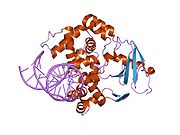
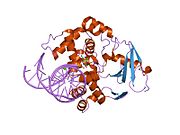
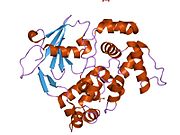
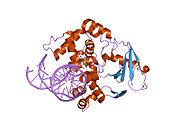
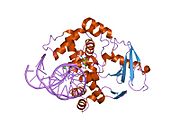
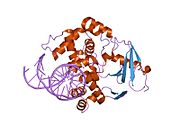
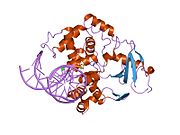
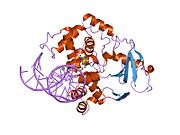
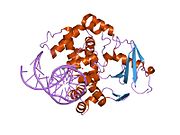
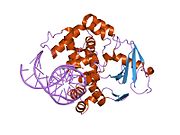
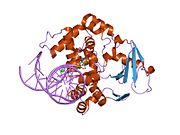
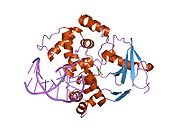
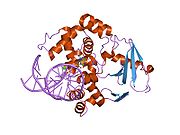
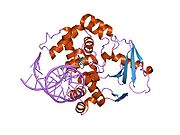
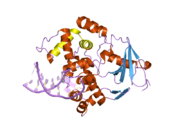
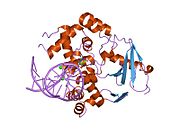
PDB gallery 1ebm: CRYSTAL STRUCTURE OF THE HUMAN 8-OXOGUANINE GLYCOSYLASE (HOGG1) BOUND TO A SUBSTRATE OLIGONUCLEOTIDE1fn7: COUPLING OF DAMAGE RECOGNITION AND CATALYSIS BY A HUMAN BASE-EXCISION DNA REPAIR PROTEIN1hu0: CRYSTAL STRUCTURE OF AN HOGG1-DNA BOROHYDRIDE TRAPPED INTERMEDIATE COMPLEX1ko9: Native Structure of the Human 8-oxoguanine DNA Glycosylase hOGG11lwv: Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-aminoguanine1lww: Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-bromoguanine1lwy: hOgg1 Borohydride-Trapped Intermediate without 8-oxoguanine1m3h: Crystal Structure of Hogg1 D268E Mutant with Product Oligonucleotide1m3q: Crystal Structure of hogg1 D268E Mutant with Base-Excised DNA and 8-aminoguanine1n39: Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase1n3a: Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase1n3c: Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase1yqk: Human 8-oxoguanine glycosylase crosslinked with guanine containing DNA1yql: Catalytically inactive hOGG1 crosslinked with 7-deaza-8-azaguanine containing DNA1yqm: Catalytically inactive human 8-oxoguanine glycosylase crosslinked to 7-deazaguanine containing DNA1yqr: Catalytically inactive human 8-oxoguanine glycosylase crosslinked to oxoG containing DNA2i5w: Structure of hOGG1 crosslinked to DNA sampling a normal G adjacent to an oxoG2nob: Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA2noe: Structure of catalytically inactive G42A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA2nof: Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA2noh: Structure of catalytically inactive Q315A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA2noi: Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA2nol: Structure of catalytically inactive human 8-oxoguanine glycosylase distal crosslink to oxoG DNA2noz: Structure of Q315F human 8-oxoguanine glycosylase distal crosslink to 8-oxoguanine DNAHydrolase: sugar hydrolases (EC 3.2) 3.2.1: Glycoside hydrolases Cellulase · Alpha-glucosidase (Acid, Neutral AB, Neutral C) · Beta-glucosidase (cytosolic) · Debranching enzymeOtherAmylase (Alpha-Amylase) · Chitinase · Lysozyme · Neuraminidase (NEU1, NEU2, NEU3, NEU4, Bacterial neuraminidase, Viral neuraminidase) · Galactosidases (Alpha, Beta) · alpha-Mannosidase · Glucuronidase · Hyaluronidase · Pullulanase · Glucosylceramidase (lysosomal, non-lysosomal) · Galactosylceramidase · Alpha-N-acetylgalactosaminidase (NAGA) · Alpha-N-acetylglucosaminidase · Fucosidase · Hexosaminidase (HEXA, HEXB) · Iduronidase · Maltase-glucoamylase · Heparanase (HPSE2)3.2.2: Hydrolysing
N-Glycosyl compoundsDNA glycosylases: Oxoguanine glycosylaseB enzm: 1.1/2/3/4/5/6/7/8/10/11/13/14/15-18, 2.1/2/3/4/5/6/7/8, 2.7.10, 2.7.11-12, 3.1/2/3/4/5/6/7, 3.1.3.48, 3.4.21/22/23/24, 4.1/2/3/4/5/6, 5.1/2/3/4/99, 6.1-3/4/5-6 Categories:- Human proteins
- Biochemistry stubs
Wikimedia Foundation. 2010.