- Purine nucleoside phosphorylase
-
Not to be confused with polynucleotide phosphorylase.
purine-nucleoside phosphorylase Identifiers EC number 2.4.2.1 CAS number 9030-21-1 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles Purine nucleoside phosphorylase also known as PNPase and inosine phosphorylase is an enzyme that in humans is encoded by the NP gene.[1]
Contents
Function
Purine nucleoside phosphorylase is an enzyme involved in purine metabolism. PNP metabolizes adenosine into adenine, inosine into hypoxanthine, and guanosine into guanine, in each case creating ribose phosphate.
Nucleoside phosphorylase is an enzyme which cleaves a nucleoside by phosphorylating the ribose to produce a nucleobase and ribose 1 phosphate. It is one enzyme of the nucleotide salvage pathways. These pathways allow the cell to produce nucleotide monophosphates when the de novo synthesis pathway has been interrupted or is non-existent (as is the case in the brain). Often the de novo pathway is interrupted as a result of chemotherapy drugs such as methotrexate or aminopterin.
All salvage pathway enzymes require a high energy phosphate donor such as ATP or PRPP.
- Thymidine can be phosphorylated by thymidine kinase (TK).
- Uridine can be phosphorylated by uridine kinase (UK).
- Cytidine can be phosphorylated by cytidine kinase (CK).
- Deoxycytidine can be phosphorylated by deoxycytidine kinase (DOK).
Adenosine uses the enzyme adenosine kinase, which is a very important enzyme in the cell. Attempts are being made to develop an inhibitor for the enzyme for use in cancer chemotherapy.
Clinical significance
PNPase together with adenosine deaminase (ADA), serves a key role in purine catabolism, referred to as the salvage pathway. Mutations in ADA lead to an accumulation of (d)ATP, which inhibits ribonucleotide reductase, leading to a deficiency in (d)CTPs and (d)TTPs, which, in turn, induces apoptosis in T-lymphocytes and B-lymphocytes, leading to severe combined immunodeficiency (SCID).[citation needed]
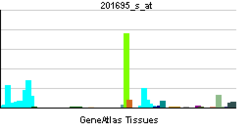
PNP-deficient patients will have an immunodeficiency problem. It affects only T-cells; B-cells are unaffected by the deficiency.
See also
References
Further reading
- Markert ML (1991). "Purine nucleoside phosphorylase deficiency.". Immunodeficiency reviews 3 (1): 45–81. PMID 1931007.
- Borgers M, Verhaegen H, De Brabander M, et al. (1978). "Purine nucleoside phosphorylase in chronic lymphocytic leukemia (CLL).". Blood 52 (5): 886–95. PMID 100152.
- Aust MR, Andrews LG, Barrett MJ, et al. (1992). "Molecular analysis of mutations in a patient with purine nucleoside phosphorylase deficiency.". Am. J. Hum. Genet. 51 (4): 763–72. PMC 1682776. PMID 1384322. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1682776.
- Andrews LG, Markert ML (1992). "Exon skipping in purine nucleoside phosphorylase mRNA processing leading to severe immunodeficiency.". J. Biol. Chem. 267 (11): 7834–8. PMID 1560016.
- Jonsson JJ, Williams SR, McIvor RS (1991). "Sequence and functional characterization of the human purine nucleoside phosphorylase promoter.". Nucleic Acids Res. 19 (18): 5015–20. doi:10.1093/nar/19.18.5015. PMC 328804. PMID 1923769. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=328804.
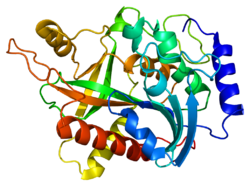
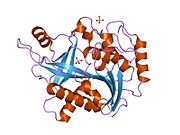
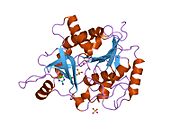
- Ealick SE, Rule SA, Carter DC, et al. (1990). "Three-dimensional structure of human erythrocytic purine nucleoside phosphorylase at 3.2 A resolution.". J. Biol. Chem. 265 (3): 1812–20. PMID 2104852.
- Williams SR, Gekeler V, McIvor RS, Martin DW (1987). "A human purine nucleoside phosphorylase deficiency caused by a single base change.". J. Biol. Chem. 262 (5): 2332–8. PMID 3029074.
- Williams SR, Goddard JM, Martin DW (1984). "Human purine nucleoside phosphorylase cDNA sequence and genomic clone characterization.". Nucleic Acids Res. 12 (14): 5779–87. doi:10.1093/nar/12.14.5779. PMC 320030. PMID 6087295. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=320030.
- Pannicke U, Tuchschmid P, Friedrich W, et al. (1997). "Two novel missense and frameshift mutations in exons 5 and 6 of the purine nucleoside phosphorylase (PNP) gene in a severe combined immunodeficiency (SCID) patient.". Hum. Genet. 98 (6): 706–9. doi:10.1007/s004390050290. PMID 8931706.
- Markert ML, Finkel BD, McLaughlin TM, et al. (1997). "Mutations in purine nucleoside phosphorylase deficiency.". Hum. Mutat. 9 (2): 118–21. doi:10.1002/(SICI)1098-1004(1997)9:2<118::AID-HUMU3>3.0.CO;2-5. PMID 9067751.
- Erion MD, Takabayashi K, Smith HB, et al. (1997). "Purine nucleoside phosphorylase. 1. Structure-function studies.". Biochemistry 36 (39): 11725–34. doi:10.1021/bi961969w. PMID 9305962.
- Erion MD, Stoeckler JD, Guida WC, et al. (1997). "Purine nucleoside phosphorylase. 2. Catalytic mechanism.". Biochemistry 36 (39): 11735–48. doi:10.1021/bi961970v. PMID 9305963.
- Stoeckler JD, Poirot AF, Smith RM, et al. (1997). "Purine nucleoside phosphorylase. 3. Reversal of purine base specificity by site-directed mutagenesis.". Biochemistry 36 (39): 11749–56. doi:10.1021/bi961971n. PMID 9305964.
- Sasaki Y, Iseki M, Yamaguchi S, et al. (1998). "Direct evidence of autosomal recessive inheritance of Arg24 to termination codon in purine nucleoside phosphorylase gene in a family with a severe combined immunodeficiency patient.". Hum. Genet. 103 (1): 81–5. doi:10.1007/s004390050787. PMID 9737781.
- Sheppard TL, Ordoukhanian P, Joyce GF (2000). "A DNA enzyme with N-glycosylase activity.". Proc. Natl. Acad. Sci. U.S.A. 97 (14): 7802–7. doi:10.1073/pnas.97.14.7802. PMC 16625. PMID 10884411. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=16625.
- Dalal I, Grunebaum E, Cohen A, Roifman CM (2001). "Two novel mutations in a purine nucleoside phosphorylase (PNP)-deficient patient.". Clin. Genet. 59 (6): 430–7. doi:10.1034/j.1399-0004.2001.590608.x. PMID 11453975.
- Ivings L, Pennington SR, Jenkins R, et al. (2002). "Identification of Ca2+-dependent binding partners for the neuronal calcium sensor protein neurocalcin delta: interaction with actin, clathrin and tubulin.". Biochem. J. 363 (Pt 3): 599–608. doi:10.1042/0264-6021:3630599. PMC 1222513. PMID 11964161. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1222513.
- Falkenberg M, Gaspari M, Rantanen A, et al. (2002). "Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA.". Nat. Genet. 31 (3): 289–94. doi:10.1038/ng909. PMID 12068295.
- Stoychev G, Kierdaszuk B, Shugar D (2002). "Xanthosine and xanthine. Substrate properties with purine nucleoside phosphorylases, and relevance to other enzyme systems.". Eur. J. Biochem. 269 (16): 4048–57. doi:10.1046/j.1432-1033.2002.03097.x. PMID 12180982.
External links
- Human PNP at Cornell University
- E. Coli PNP at Cornell University
- MeSH Purine-Nucleoside+Phosphorylase
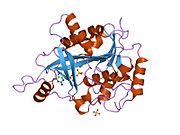
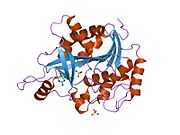
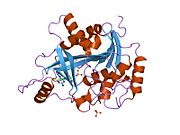
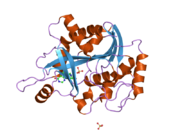
PDB gallery 1m73: CRYSTAL STRUCTURE OF HUMAN PNP AT 2.3A RESOLUTION1pf7: CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH IMMUCILLIN H1pwy: CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH ACYCLOVIR1rct: Crystal structure of Human purine nucleoside phosphorylase complexed with INOSINE1rfg: Crystal Structure of Human Purine Nucleoside Phosphorylase Complexed with Guanosine1rr6: Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and phosphate1rsz: Structure of human purine nucleoside phosphorylase in complex with DADMe-Immucillin-H and sulfate1rt9: Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and sulfate1ula: APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS1ulb: APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS1v2h: Crystal structure of human PNP complexed with guanine1v3q: Structure of human PNP complexed with DDI1v41: Crystal structure of human PNP complexed with 8-Azaguanine1v45: Crystal Structure of human PNP complexed with 3-deoxyguanosine1yry: Crystal structure of human PNP complexed with MESG2a0w: Structure of human purine nucleoside phosphorylase H257G mutant2a0x: Structure of human purine nucleoside phosphorylase H257F mutant2a0y: Structure of human purine nucleoside phosphorylase H257D mutant2oc4: Crystal stucture of human purine nucleoside phosphorylase mutant H257D with Imm-H2oc9: Crystal stucture of human purine nucleoside phosphorylase mutant H257G with Imm-H2on6: Crystal stucture of human purine nucleoside phosphorylase mutant H257F with Imm-HTransferases: glycosyltransferases (EC 2.4) 2.4.1: Hexosyl-
transferases2.4.2: Pentosyl-
transferasesOtherPurine nucleoside phosphorylase: Thymidine phosphorylase (ECGF1)Other2.4.99: Sialyl
transferasesPurine metabolism AnabolismR5P->IMP: Ribose-phosphate diphosphokinase · Amidophosphoribosyltransferase · Phosphoribosylglycinamide formyltransferase · AIR synthetase (FGAM cyclase) · Phosphoribosylaminoimidazole carboxylase · Phosphoribosylaminoimidazolesuccinocarboxamide synthase · IMP synthase
IMP->AMP: Adenylosuccinate synthase · Adenylosuccinate lyase · reverse (AMP deaminase)
IMP->GMP: IMP dehydrogenase · GMP synthase · reverse (GMP reductase)CatabolismAdenosine deaminase · Purine nucleoside phosphorylase · Guanine deaminase · Xanthine oxidase · Urate oxidasePyrimidine metabolism AnabolismCatabolismDeoxyribonucleotides This transferase article is a stub. You can help Wikipedia by expanding it.