- AMP deaminase
-
Adenosine monophosphate deaminase 1 Identifiers Symbols AMPD1; MAD; MADA External IDs OMIM: 102770 MGI: 88015 HomoloGene: 20 GeneCards: AMPD1 Gene EC number 3.5.4.6 Gene Ontology Molecular function • AMP deaminase activity
• hydrolase activity
• metal ion bindingCellular component • cytosol Biological process • purine base metabolic process
• nucleotide metabolic process
• purine ribonucleoside monophosphate biosynthetic process
• response to organic substance
• purine-containing compound salvage
• nucleobase, nucleoside and nucleotide metabolic processSources: Amigo / QuickGO RNA expression pattern 
More reference expression data Orthologs Species Human Mouse Entrez 270 229665 Ensembl ENSG00000116748 ENSMUSG00000070385 UniProt P23109 Q3V1D3 RefSeq (mRNA) NM_000036.2 NM_001033303.2 RefSeq (protein) NP_000027.2 NP_001028475.2 Location (UCSC) Chr 1:
115.22 – 115.24 MbChr 3:
102.88 – 102.9 MbPubMed search [1] [2] AMP deaminase 1 is an enzyme that in humans is encoded by the AMPD1 gene.[1][2]
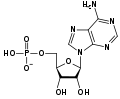
Adenosine monophosphate deaminase is an enzyme that converts adenosine monophosphate (AMP) to inosine monophosphate (IMP), freeing an ammonia molecule in the process.
Adenosine monophosphate deaminase 1 catalyzes the deamination of AMP to IMP in skeletal muscle and plays an important role in the purine nucleotide cycle. Two other genes have been identified, AMPD2 and AMPD3, for the liver- and erythrocyte-specific isoforms, respectively. Deficiency of the muscle-specific enzyme is apparently a common cause of exercise-induced myopathy and probably the most common cause of metabolic myopathy in the human.[2]
Contents
Function
It is a part of the metabolic process that converts sugar, fat, and protein into cellular energy.
In order to use energy, a cell converts one of the above fuels into adenosine triphosphate (ATP) via the mitochondria. Cellular processes, especially muscles, then convert the ATP into adenosine diphosphate (ADP), freeing the energy to do work.
A new research report [3] shows that the widely-prescribed diabetes medication metformin http://en.wikipedia.org/wiki/Metformin works on AMP kinase by directly inhibiting AMP deaminase, thereby increasing cellular AMP.
Pathology
A deficiency is associated with myoadenylate deaminase deficiency.
References
- ^ Mahnke-Zizelman DK, Sabina RL (Nov 1992). "Cloning of human AMP deaminase isoform E cDNAs. Evidence for a third AMPD gene exhibiting alternatively spliced 5'-exons". J Biol Chem 267 (29): 20866–77. PMID 1400401.
- ^ a b "Entrez Gene: AMPD1 adenosine monophosphate deaminase 1 (isoform M)". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=270.
- ^ http://www.ncbi.nlm.nih.gov/pubmed/21059655
Further reading
- Fishbein WN, Armbrustmacher VW, Griffin JL (1978). "Myoadenylate deaminase deficiency: a new disease of muscle.". Science 200 (4341): 545–8. doi:10.1126/science.644316. PMID 644316.
- Sabina RL, Fishbein WN, Pezeshkpour G, et al. (1992). "Molecular analysis of the myoadenylate deaminase deficiencies.". Neurology 42 (1): 170–9. PMID 1370861.
- Morisaki T, Gross M, Morisaki H, et al. (1992). "Molecular basis of AMP deaminase deficiency in skeletal muscle.". Proc. Natl. Acad. Sci. U.S.A. 89 (14): 6457–61. doi:10.1073/pnas.89.14.6457. PMC 49520. PMID 1631143. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=49520.
- Sabina RL, Morisaki T, Clarke P, et al. (1990). "Characterization of the human and rat myoadenylate deaminase genes.". J. Biol. Chem. 265 (16): 9423–33. PMID 2345176.
- Dale GL (1989). "Radioisotopic assay for erythrocyte adenosine 5'-monophosphate deaminase.". Clin. Chim. Acta 182 (1): 1–7. doi:10.1016/0009-8981(89)90144-7. PMID 2502331.
- Mercelis R, Martin JJ, Dehaene I, et al. (1981). "Myoadenylate deaminase deficiency in a patient with facial and limb girdle myopathy.". J. Neurol. 225 (3): 157–66. doi:10.1007/BF00313744. PMID 6167680.
- van Laarhoven JP, de Gast GC, Spierenburg GT, de Bruyn CH (1983). "Enzymological studies in chronic lymphocytic leukemia.". Leuk. Res. 7 (2): 261–7. doi:10.1016/0145-2126(83)90016-4. PMID 6406772.
- Kelemen J, Rice DR, Bradley WG, et al. (1982). "Familial myoadenylate deaminase deficiency and exertional myalgia.". Neurology 32 (8): 857–63. PMID 7201581.
- Baumeister FA, Gross M, Wagner DR, et al. (1993). "Myoadenylate deaminase deficiency with severe rhabdomyolysis.". Eur. J. Pediatr. 152 (6): 513–5. doi:10.1007/BF01955062. PMID 8335021.
- Morisaki T, Holmes EW (1993). "Functionally distinct elements are required for expression of the AMPD1 gene in myocytes.". Mol. Cell. Biol. 13 (9): 5854–60. PMC 360332. PMID 8355716. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=360332.
- Bruno C, Minetti C, Shanske S, et al. (1998). "Combined defects of muscle phosphofructokinase and AMP deaminase in a child with myoglobinuria.". Neurology 50 (1): 296–8. PMID 9443500.
- Hisatome I, Morisaki T, Kamma H, et al. (1998). "Control of AMP deaminase 1 binding to myosin heavy chain.". Am. J. Physiol. 275 (3 Pt 1): C870–81. PMID 9730972.
- Sims B, Powers RE, Sabina RL, Theibert AB (1999). "Elevated adenosine monophosphate deaminase activity in Alzheimer's disease brain.". Neurobiol. Aging 19 (5): 385–91. doi:10.1016/S0197-4580(98)00083-9. PMID 9880040.
- Loh E, Rebbeck TR, Mahoney PD, et al. (1999). "Common variant in AMPD1 gene predicts improved clinical outcome in patients with heart failure.". Circulation 99 (11): 1422–5. PMID 10086964.
- Abe M, Higuchi I, Morisaki H, et al. (2000). "Myoadenylate deaminase deficiency with progressive muscle weakness and atrophy caused by new missense mutations in AMPD1 gene: case report in a Japanese patient.". Neuromuscul. Disord. 10 (7): 472–7. doi:10.1016/S0960-8966(00)00127-9. PMID 10996775.
- Morisaki H, Higuchi I, Abe M, et al. (2001). "First missense mutations (R388W and R425H) of AMPD1 accompanied with myopathy found in a Japanese patient.". Hum. Mutat. 16 (6): 467–72. doi:10.1002/1098-1004(200012)16:6<467::AID-HUMU3>3.0.CO;2-V. PMID 11102975.
- Gross M, Rötzer E, Kölle P, et al. (2002). "A G468-T AMPD1 mutant allele contributes to the high incidence of myoadenylate deaminase deficiency in the Caucasian population.". Neuromuscul. Disord. 12 (6): 558–65. doi:10.1016/S0960-8966(02)00008-1. PMID 12117480.
- Mahnke-Zizelman DK, Sabina RL (2003). "N-terminal sequence and distal histidine residues are responsible for pH-regulated cytoplasmic membrane binding of human AMP deaminase isoform E.". J. Biol. Chem. 277 (45): 42654–62. doi:10.1074/jbc.M203473200. PMID 12213808.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=139241.
-
adenosine monophosphate (AMP)
External links
Purine metabolism AnabolismR5P->IMP: Ribose-phosphate diphosphokinase · Amidophosphoribosyltransferase · Phosphoribosylglycinamide formyltransferase · AIR synthetase (FGAM cyclase) · Phosphoribosylaminoimidazole carboxylase · Phosphoribosylaminoimidazolesuccinocarboxamide synthase · IMP synthase
IMP->AMP: Adenylosuccinate synthase · Adenylosuccinate lyase · reverse (AMP deaminase)
IMP->GMP: IMP dehydrogenase · GMP synthase · reverse (GMP reductase)CatabolismPyrimidine metabolism AnabolismCatabolismDeoxyribonucleotides Categories:- Human proteins
- Hydrolase stubs
Wikimedia Foundation. 2010.