- Escherichia coli O157:H7
-
Escherichia coli O157:H7 Classification and external resources ICD-10 A04.3 ICD-9 008.04 Escherichia coli O157:H7 is an enterohemorrhagic strain of the bacterium Escherichia coli and a cause of foodborne illness.[1] Infection often leads to hemorrhagic diarrhea, and occasionally to kidney failure, especially in young children and elderly persons. Transmission is via the fecal-oral route, and most illness has been associated with eating undercooked, contaminated ground beef, swimming in or drinking contaminated water, and eating contaminated vegetables.
Contents
Bacteriology
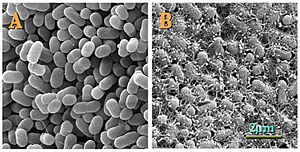
 E. coli O157:H7
E. coli O157:H7
E. coli serotype O157:H7 is a Gram-negative, rod-shaped bacterium. The "O" in the name refers to the cell wall (somatic) antigen number, whereas the "H" refers to the flagella antigen. Other serotypes may cause (usually less severe) illness, but only those with the specific O157:H7 combination are reviewed here. Other bacteria may be classified by "K" or capsular antigens. (The "O" stands for ohne Hauch [Ger. "without huff" or "without film"]; "H" for Hauch; and "K" for Kapsel.[2][3]) This is one of hundreds of serotypes of the bacterium Escherichia coli. While most strains are harmless and normally found in the intestines of mammals, this strain may produce shiga-like toxins and cause severe illness, and is a member of a class of pathogenic E. coli known as enterohemorrhagic Escherichia coli or EHEC. Often they are referred to by their toxin producing capabilities, verocytotoxin producing E. coli (VTEC) or Shiga-like toxin producing E. coli (STEC).
E. coli O157:H7 was first recognized as a pathogen as a result of an outbreak of unusual gastrointestinal illness in 1982. The outbreak was traced to contaminated hamburgers, and the illness was similar to other incidents in the United States and Japan. The etiologic agent of the illness was identified as a rare O157:H7 serotype of E. coli in 1983. This serotype had only been isolated once before, from a sick patient in 1975.[4]
E. coli O157:H7 is markedly different from other pathogenic E. coli, as well. In particular, the O157:H7 serotype is negative for invasiveness (Sereny test), adheres through the E. coli common pilus (ECP),[5] and does not produce heat stable or heat labile toxins. In addition, E. coli O157:H7 is usually sorbitol negative, whereas 93% of all E. coli ferment sorbitol.[6] E. coli O157:H7 also lacks the ability to hydrolyze 4-methylumbelliferyl-β-D-glucuronide (MUG)[7] and does not grow at 45 °C in the presence of 0.15% bile salts. Because of the latter characteristic, this serotype cannot be isolated by using standard fecal coliform methods that include incubation at 45 °C.[7][8]
E. coli O157:H7 serotypes are closely related, descended from a common ancestor, divergent in plasmid content more than chromosomal content, and are no more related to other shiga toxin producing strains than any other randomly chosen E. coli serotype. E. coli O55:H7 and E. coli O157:H7 are most closely related and diverged from a common pathogenic ancestor that possessed the ability to form attaching and effacing lesions. E. coli O157:H7 serotypes apparently arose as a result of horizontal gene transfer of virulence factors.[9]
Among these virulence factors are a periplasmic catalase and shiga-like toxins. Shiga-like toxins are iron-regulated toxins that catalytically inactivate 60S ribosomal subunits of eukaryotic cells, blocking mRNA translation and causing cell death.[10] Shiga-like toxins are functionally identical to toxins produced by virulent Shigella species.[11] Strains of E. coli that express shiga-like toxins gained this ability due to infection with a prophage containing the structural coding for the toxin, and nonproducing strains may become infected and produce shiga-like toxins after incubation with shiga toxin positive strains. The prophage responsible seems to have infected the strain's ancestors fairly recently, as viral particles have been observed to replicate in the host if it is stressed in some way (e.g. antibiotics).[12][13] The periplasmic catalase is encoded on the pO157 plasmid, and is believed to be involved in virulence by providing additional oxidative protection when infecting the host.[14]
Natural Habitat
While relatively uncommon, E. coli O157:H7 can naturally be found in the intestinal contents of some cattle.[15] Because ruminants lack a receptor for the toxin the bacteria produce, it does not cause disease in them and is considered commensal.
Transmission
Infection with E. coli O157:H7 follows ingestion of contaminated food or water, or oral contact with contaminated surfaces. It is highly virulent, with a low infectious dose: an inoculation of fewer than 10 to 100 CFU of E. coli O157:H7 is sufficient to cause infection, compared to over one-million CFU for other pathogenic E. coli strains.[16]
A main source of infection is undercooked ground beef; other sources include consumption of unpasteurized milk and juice, raw produce and salami, and contact with infected live animals. Waterborne transmission occurs through swimming in contaminated lakes, pools, or drinking inadequately treated water. The organism is easily transmitted from person to person and has been difficult to control in child day-care centers.
E. coli O157:H7 is found on cattle farms and can live in the intestines of healthy cattle. The toxin requires highly specific receptors on the cells' surface in order to attach and enter the cell; species such as cattle, swine, and deer which do not carry these receptors may harbor toxigenic bacteria without any ill effect, shedding them in their feces, from which they may be spread to humans. Flesh can become contaminated during slaughter and butchering, and organisms can be thoroughly mixed into beef when it is ground into hamburger. Bacteria present on the cow's udders or on equipment may get into raw milk. Contaminated foods look, smell, and taste the same as their normal counterparts.
Another potential vector of E. coli O157:H7 is filth flies (which includes house flies, Musca domestica). Filth flies have been shown to be vectors of E. coli O157:H7 using PCR.[17][18]
United States food advocates have unsuccessfully attempted to control the spread of this illness by promoting the so-called "Kevin's Law". This law would give the United States Department of Agriculture power to shut down food processing plants that fail multiple inspections. The food processing industry vigorously opposes this proposal.[19]
Signs and symptoms
E. coli O157:H7 infection often causes severe, acute hemorrhagic diarrhea (although nonhemorrhagic diarrhea is also possible) and abdominal cramps. Usually little or no fever is present, and the illness resolves in five to 10 days. It can also be asymptomatic.[citation needed]
In some people, particularly children under five years of age and the elderly, the infection can cause hemolytic uremic syndrome (HUS), in which the red blood cells are destroyed and the kidneys fail. About 2–7% of infections lead to this complication. In the United States, HUS is the principal cause of acute kidney failure in children, and most cases of HUS are caused by E. coli O157:H7.
Diagnosis
A stool culture can detect the bacterium, although it is not a routine test and so must be specifically requested. The sample is cultured on sorbitol-MacConkey (SMAC) agar, or the variant cefixime potassium tellurite sorbitol-MacConkey agar (CT-SMAC). On SMAC agar O157 colonies appear clear due to their inability to ferment sorbitol, while the colonies of the usual sorbitol-fermenting serotypes of E. coli appear red. Sorbitol nonfermenting colonies are tested for the somatic O157 antigen before being confirmed as E. coli O157. Like all cultures, diagnosis is time-consuming with this method; swifter diagnosis is possible using PCR techniques. Newer technologies using fluorescent and antibody detection are also under development.
Surveillance
E. coli O157:H7 infection is nationally reportable in the USA and Great Britain, and is reportable in most US states.
Treatment
While fluid replacement and blood pressure support may be necessary to prevent death from dehydration, most victims recover without treatment in five to 10 days. There is no evidence that antibiotics improve the course of disease, and treatment with antibiotics may precipitate kidney complications.[20] Antidiarrheal agents, such as loperamide (imodium), should also be avoided.
Hemolytic-uremic syndrome is a life-threatening condition usually treated in an intensive care unit. Blood transfusions and kidney dialysis are often required. With intensive care, the death rate for hemolytic uremic syndrome is 3–5%.
Costs
The pathogen results in an estimated 2,100 hospitalizations annually in the United States. The illness is often misdiagnosed; therefore, expensive and invasive diagnostic procedures may be performed. Patients who develop HUS often require prolonged hospitalization, dialysis, and long-term followup.
Prevention
Agricultural
Beef processing is a common point of contamination: during the slaughtering process, the contents of intestines or fecal material on the hide could mix with the meat, thus allowing bacteria to flourish in the warm, damp conditions. If the infected parts are then ground, the bacteria go from the surface of the cut to the interior of the ground mass. Ground beef is therefore much more likely to be a source of infection than steak. In steak, only the surface area of a cut is exposed during butchering, and cooking the outside affects the entire exposed portion. In ground beef, however, bacteria are mixed throughout the meat mass, requiring the entire mass to be heated thoroughly to eliminate the pathogen. Additionally, in the production of ground beef, meat from multiple cattle is often ground together, enabling contamination from a single animal to contaminate an entire lot of ground beef.
Accordingly, reduction of infection requires preventive measures that either reduce the number of cattle that carry E. coli O157:H7 or reduce the contamination of meat during slaughter and grinding. These measures include careful removal and cleaning of the intestines, steam/vacuum treatment, and organic acid sprays.
In January 2007, Canadian biopharmaceutical company Bioniche announced it had developed a bovine vaccine capable of reducing O157:H7 in cattle by over 99%.[21]
On March 4, 2009, the United States Department of Agriculture (USDA) granted Epitopix a conditional license for the first E. coli O157 vaccine for cattle in the U.S.[22]
In May 2010, a paper from the University of Texas Southwestern Medical Center detailed preliminary results of research into the sensing mechanisms used by O157:H7 to determine when to adopt an acid-resistant state needed to escape from the digestive tract. The study introduced two different strains of E. coli lacking normal sensing mechanisms into the rumens of grain-fed cattle. The strains were unable to pass through the stomach. Next, the researchers plan to conduct tests on grass-fed cattle to establish if the mechanism is the same.[23][24]
Culinary and dietary
Cooking all ground beef and hamburgers thoroughly, and checking the temperature using an instant-read meat thermometer, will eliminate the organism. Ground beef should be cooked until a thermometer inserted into several parts of the patty, including the thickest part, reads at least 72 °C (162 °F).
When preparing meat, it should be kept separate from other food items and all surfaces, and utensils which come into contact with raw meat should be washed thoroughly before being used again. Hand washing is similarly important. Placing cooked hamburgers or ground beef on an unwashed plate that held raw patties can transmit infection. This was the main cause of the E. coli poisoning in Scotland in 1996 that killed seven and left hundreds infected.
Unpasteurized milk and juices are potential sources of E. coli. Commercial juice is almost always pasteurized, and juice concentrates are also heated sufficiently to kill pathogens.
Fruits and vegetables should be washed thoroughly, especially those that will not be cooked. Children under five years of age, immunocompromised persons, and the elderly should avoid eating alfalfa sprouts until their safety can be assured. Methods to decontaminate alfalfa seeds and sprouts are being investigated.
Before consumption, contaminated water should be sterilized by 1) boiling at a rolling boil for at least one minute (longer at higher altitudes), 2) ultrafiltration using a system designed for water treatment, or 3) chemically sterilized using an ozone, chlorine, or iodine product. Care while swimming to avoid ingestion of potentially contaminated water can reduce the chances of infection.
Proper hand washing after using the lavatory or changing a diaper, especially among children or those with diarrhea, reduces the risk of transmission. Anyone with a diarrheal illness should avoid swimming in public pools or lakes, sharing baths with others, and preparing food for others.
United States
The U.S.D.A. banned the sale of ground beef contaiminated with the O157:H7 strain in 1994.[25]
See also
- 2011 E. coli O104:H4 outbreak
- Centers for Disease Control and Prevention
- Escherichia coli O104:H4
- Escherichia coli O121
- Food-induced purpura
- List of foodborne illness outbreaks
References
- ^ Karch H, Tarr P, Bielaszewska M (2005). "Enterohaemorrhagic Escherichia coli in human medicine.". Int J Med Microbiol 295 (6–7): 405–18. doi:10.1016/j.ijmm.2005.06.009. PMID 16238016.
- ^ See Kauffmann-White-Schema in the German Wikipedia.
- ^ O antigen at Dorlands Medical Dictionary.
- ^ Riley L, Remis R, Helgerson S, McGee H, Wells J, Davis B, Hebert R, Olcott E, Johnson L, Hargrett N, Blake P, Cohen M (1983). "Hemorrhagic colitis associated with a rare Escherichia coli serotype". N Engl J Med 308 (12): 681–5. doi:10.1056/NEJM198303243081203. PMID 6338386.
- ^ Rendón, M. A. et al. (2007). "Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization". PNAS 104 (25): 10637–10642. doi:10.1073/pnas.0704104104. PMC 1890562. PMID 17563352. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1890562.
- ^ Wells JG, Davis BR, Wachsmuth IK et al. (September 1983). "Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype". Journal of clinical microbiology 18 (3): 512–20. PMC 270845. PMID 6355145. http://jcm.asm.org/cgi/pmidlookup?view=long&pmid=6355145.
- ^ a b Doyle MP, Schoeni JL (October 1984). "Survival and growth characteristics of Escherichia coli associated with hemorrhagic colitis". Applied and environmental microbiology 48 (4): 855–6. PMC 241626. PMID 6391379. http://aem.asm.org/cgi/pmidlookup?view=long&pmid=6391379.
- ^ Szabo, R. A.; Todd, E. C. D.; Jean, A. (1986). "Method to isolate Escherichia coli 0157:H7 from food". Journal of Food Protection 49 (10): 768–772. ISSN 0362-028X.
- ^ Whittam TS, Wachsmuth IK, Wilson RA (June 1988). "Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome". The Journal of infectious diseases 157 (6): 1124–33. doi:10.1093/infdis/157.6.1124. PMID 3286782.
- ^ Reisbig R, Olsnes S, Eiklid K (August 1981). "The cytotoxic activity of Shigella toxin. Evidence for catalytic inactivation of the 60 S ribosomal subunit". The Journal of biological chemistry 256 (16): 8739–44. PMID 6894922. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=6894922.
- ^ Calderwood SB, Auclair F, Donohue-Rolfe A, Keusch GT, Mekalanos JJ (July 1987). "Nucleotide sequence of the Shiga-like toxin genes of Escherichia coli". Proceedings of the National Academy of Sciences of the United States of America 84 (13): 4364–8. doi:10.1073/pnas.84.13.4364. PMC 305089. PMID 3299365. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=3299365.
- ^ O'Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB (November 1984). "Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea". Science 226 (4675): 694–6. doi:10.1126/science.6387911. PMID 6387911.
- ^ Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O'Brien AD (July 1986). "Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities". Infection and immunity 53 (1): 135–40. PMC 260087. PMID 3522426. http://iai.asm.org/cgi/pmidlookup?view=long&pmid=3522426.
- ^ Brunder W, Schmidt H, Karch H (November 1996). "KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7". Microbiology (Reading, England) 142 (Pt 11): 3305–15. PMID 8969527.
- ^ "Escherichia coli". http://science.jrank.org/pages/2571/Escherichia-coli.html. Retrieved June 24, 2010.
- ^ J.D. Greig, E.C.D. Todd, C. Bartleson, and B. Michaels. March 25, 2010. "Infective Doses and Pathen Carriage", pp. 19-20, USDA 2010 Food Safety Education Conference.
- ^ Brazil, S. M.; Steelman, C. D.; Szalanski, A. L. (2007). "Detection of pathogen DNA from filth flies (Diptera: Muscidae) using filter paper spot cards". Journal of Agricultural and Urban Entomology 24 (1): 13–18. doi:10.3954/1523-5475-24.1.13.
- ^ Szalanski, A. L.; Owens, C. B.; McKay, T.; Steelman, C. D. (2004). "Detection of Campylobacter sp., and E. coli O157:H7 in filth flies (Diptera: Muscidae) by polymerase chain reaction". Medical and Veterinary Entomology 18 (3): 241–246. doi:10.1111/j.0269-283X.2004.00502.x. PMID 15347391. http://comp.uark.edu/~aszalan/med_vet_ent_2004.pdf.
- ^ Food Inc. Directed by Robert Kenner, Independent Films 2009
- ^ Walterspiel JN, Ashkenazi S, Morrow AL, Cleary TG (1992). "Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin I". Infection 20 (1): 25–9. doi:10.1007/BF01704889. PMID 1563808.
- ^ "Canadian Research Collaboration Produces World's First Food Safety Vaccine: Against E. coli 0157:H7". CNX Marketlink. http://www.cnxmarketlink.com/en/releases/archive/January2007/10/c4698.html. Retrieved 2007-09-15.
- ^ Epitopix.com
- ^ UT Southwestern Medical Center (12 May 2010). "Mechanism That May Stop E. Coli from Developing in Cattle Identified". ScienceDaily. http://www.sciencedaily.com/releases/2010/05/100511111931.htm. Retrieved 13 May 2010. Full paper in Proceedings of the National Academy of Sciences
- ^ Hughes, David T.; Terekhova, D. A. et al. (2010). "Chemical sensing in mammalian host-bacterial commensal associations". PNAS 107 (21): 9831–9836. doi:10.1073/pnas.1002551107.
- ^ "Ban on E. Coli in Ground Beef Is to Extend to 6 More Strains". New York Times. September 12, 2011. http://www.nytimes.com/2011/09/13/business/federal-officials-extend-e-coli-ban.html?scp=2&sq=Jack%20in%20the%20Box&st=cse. Retrieved 2011-10-08. "After the U.S.D.A. banned the O157 form of E. coli from ground beef in 1994, the meat industry sued to block the move, but the agency prevailed in court."
External links
- E. coli: Protecting yourself and your family from a sometimes deadly bacterium
- Escherichia coli O157:H7 genomes and related information at PATRIC, a Bioinformatics Resource Center funded by NIAID
- For more information about reducing your risk of foodborne illness, visit the US Department of Agriculture's Food Safety and Inspection Service website or the Partnership for Food Safety Education
- Cooking Ground Beef Safely
- briandeer.com, report from The Sunday Times on a UK outbreak, May 17, 1998
- CBS5 report on September 2006 outbreak
Consumer food safety Adulterants/food contaminants 3-MCPD · Aldicarb · Cyanide · Formaldehyde · Lead poisoning · Melamine · Mercury in fish · Sudan red dye
Toxins/poisons Aflatoxin · Arsenic contamination of groundwater · Benzene in soft drinks · Bisphenol A · Mycotoxins · Shellfish poisoning
Microorganisms Botulism · Campylobacter jejuni · Clostridium perfringens · Escherichia coli O104:H4 · Escherichia coli O157:H7 · Hepatitis A · Hepatitis E · Listeria · Norovirus · Rotavirus · Salmonella
Pesticides overuse/residues Chlorpyrifos · DDT · Lindane · Malathion · Methamidophos
Preservatives Sweeteners Acesulfame potassium · Aspartame · Cyclamate · High-fructose corn syrup (Health effects, Public relations) · Saccharin · Sorbitol · Sucralose
Food scares 2005 Indonesia food scare · 2006 North American E. coli outbreak · 2007 Vietnam food scare · 2008 Canadian listeriosis outbreak · 2008 Chinese milk scandal · 2008 Irish pork crisis · 2008 United States salmonellosis outbreak · 2011 E. coli O104:H4 outbreak · Food safety incidents in the People's Republic of China · Bradford sweets poisoning · Chilean grape scare · ICA meat repackaging controversy · Minamata disease · Toxic oil syndrome · List of foodborne illness outbreaks · List of food contamination incidents
Regulatory/watchdog Acceptable daily intake · E number · Early history of food regulation in the United States) · European Food Safety Authority · Food and Drug Administration · Food labeling regulations · (United Kingdom and United States Food law · Food Safety Act 1990 · Food Standards Agency · International Food Safety Network · Pure Food and Drug Act · Quality Assurance International · List of food safety organisations
Food processing 4-Hydroxynonenal · Acrylamide · Creutzfeldt–Jakob disease · Food irradiation · Heterocyclic amines · Hydrolyzed vegetable protein · Modified starch · Nitrosamines · Polycyclic aromatic hydrocarbon · Shortening · Trans fat
Miscellaneous Categories:- Escherichia coli
- Foodborne illnesses
Wikimedia Foundation. 2010.