- Plasmid
-
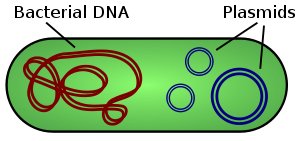
In microbiology and genetics, a plasmid is a DNA molecule that is separate from, and can replicate independently of, the chromosomal DNA.[1] They are double-stranded and, in many cases, circular. Plasmids usually occur naturally in bacteria, but are sometimes found in eukaryotic organisms (e.g., the 2-micrometre ring in Saccharomyces cerevisiae).
Plasmid sizes vary from 1 to over 1,000 kbp. The number of identical plasmids in a single cell can range anywhere from one to even thousands under some circumstances. Plasmids can be considered part of the mobilome because they are often associated with conjugation, a mechanism of horizontal gene transfer.
The term plasmid was first introduced by the American molecular biologist Joshua Lederberg in 1952.[2]
Plasmids are considered "replicons", capable of autonomous replication within a suitable host. Plasmids can be found in all three major domains: Archea, Bacteria, and Eukarya.[1] Similar to viruses, plasmids are not considered by some to be a form of "life".[3] Unlike viruses, plasmids are "naked" DNA and do not encode genes necessary to encase the genetic material for transfer to a new host, though some classes of plasmids encode the sex pilus necessary for their own transfer. Plasmid host-to-host transfer requires direct, mechanical transfer by conjugation or changes in host gene expression allowing the intentional uptake of the genetic element by transformation.[1] Microbial transformation with plasmid DNA is neither parasitic nor symbiotic in nature, because each implies the presence of an independent species living in a commensal or detrimental state with the host organism. Rather, plasmids provide a mechanism for horizontal gene transfer within a population of microbes and typically provide a selective advantage under a given environmental state. Plasmids may carry genes that provide resistance to naturally occurring antibiotics in a competitive environmental niche, or the proteins produced may act as toxins under similar circumstances. Plasmids can also provide bacteria with the ability to fix elemental nitrogen or to degrade recalcitrant organic compounds that provide an advantage when nutrients are scarce.[1]
Contents
Vectors
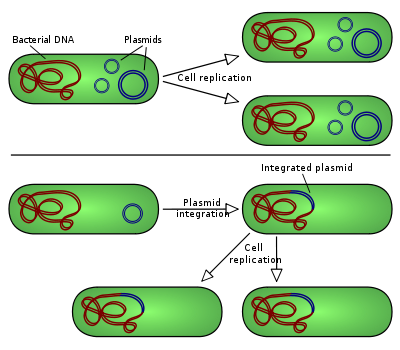 There are two types of plasmid integration into a host bacteria: Non-integrating plasmids replicate as with the top instance, whereas episomes, the lower example, integrate into the host chromosome.
There are two types of plasmid integration into a host bacteria: Non-integrating plasmids replicate as with the top instance, whereas episomes, the lower example, integrate into the host chromosome. Further information: Vector (molecular biology)
Further information: Vector (molecular biology)Plasmids used in genetic engineering are called vectors. Plasmids serve as important tools in genetics and biotechnology labs, where they are commonly used to multiply (make many copies of) or express particular genes.[4] Many plasmids are commercially available for such uses. The gene to be replicated is inserted into copies of a plasmid containing genes that make cells resistant to particular antibiotics and a multiple cloning site (MCS, or polylinker), which is a short region containing several commonly used restriction sites allowing the easy insertion of DNA fragments at this location. Next, the plasmids are inserted into bacteria by a process called transformation. Then, the bacteria are exposed to the particular antibiotics. Only bacteria that take up copies of the plasmid survive, since the plasmid makes them resistant. In particular, the protecting genes are expressed (used to make a protein) and the expressed protein breaks down the antibiotics. In this way, the antibiotics act as a filter to select only the modified bacteria. Now these bacteria can be grown in large amounts, harvested, and lysed (often using the alkaline lysis method) to isolate the plasmid of interest.
Another major use of plasmids is to make large amounts of proteins. In this case, researchers grow bacteria containing a plasmid harboring the gene of interest. Just as the bacterium produces proteins to confer its antibiotic resistance, it can also be induced to produce large amounts of proteins from the inserted gene. This is a cheap and easy way of mass-producing a gene or the protein it then codes for, for example, insulin or even antibiotics.
However, a plasmid can contain inserts of only about 1–10 kbp. To clone longer lengths of DNA, lambda phage with lysogeny genes deleted, cosmids, bacterial artificial chromosomes, or yeast artificial chromosomes are used.
Applications
Disease models
Plasmids were historically used to genetically engineer the embryonic stem cells of rats in order to create rat genetic disease models. The limited efficiency of plasmid-based techniques precluded their use in the creation of more accurate human cell models. However, developments in Adeno-associated virus recombination techniques, and Zinc finger nucleases, have enabled the creation of a new generation of isogenic human disease models.
Gene therapy
The success of some strategies of gene therapy depends on the efficient insertion of therapeutic genes at the appropriate chromosomal target sites within the human genome, without causing cell injury, oncogenic mutations (cancer) or an immune response. Plasmid vectors are one of many approaches that could be used for this purpose. Zinc finger nucleases (ZFNs) offer a way to cause a site-specific double-strand break to the DNA genome and cause homologous recombination. This makes targeted gene correction a possibility in human cells. Plasmids encoding ZFN could be used to deliver a therapeutic gene to a pre-selected chromosomal site with a frequency higher than that of random integration. Although the practicality of this approach to gene therapy has yet to be proven, some aspects of it could be less problematic than the alternative viral-based delivery of therapeutic genes.[5]
Episomes
Episomes are the eukaryotic equivalent of bacterial plasmids. In general, in eukaryotes, episomes are closed circular DNA molecules that are replicated in the nucleus. Viruses are the most common examples of this, such as herpesviruses, adenoviruses, and polyomaviruses. Other examples include aberrant chromosomal fragments, such as double minute chromosomes, that can arise during artificial gene amplifications or in pathologic processes (e.g., cancer cell transformation). Episomes in eukaryotes behave similarly to plasmids in prokaryotes in that the DNA is stably maintained and replicated with the host cell. Cytoplasmic viral episomes (as in poxvirus infections) can also occur. Some episomes, such as herpesviruses, replicate in a rolling circle mechanism, similar to bacterial phage viruses. Others replicate through a bidirectional replication mechanism. In either case, episomes remain physically separate from host cell chromosomes. Several cancer viruses, including Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus, are maintained as latent, chromosomally-distinct episomes in cancer cells, where the viruses express oncogenes that promote cancer cell proliferation. In cancers, these episomes passively replicate together with host chromosomes when the cell divides. When these viral episomes initiate lytic replication to generate multiple virus particles, they in general activate cellular innate immunity defense mechanisms that kill the host cell.
Types
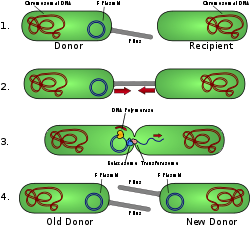
One way of grouping plasmids is by their ability to transfer to other bacteria. Conjugative plasmids contain tra genes, which perform the complex process of conjugation, the transfer of plasmids to another bacterium (Fig. 4). Non-conjugative plasmids are incapable of initiating conjugation, hence they can be transferred only with the assistance of conjugative plasmids. An intermediate class of plasmids are mobilizable, and carry only a subset of the genes required for transfer. They can parasitize a conjugative plasmid, transferring at high frequency only in its presence. Plasmids are now being used to manipulate DNA and may possibly be a tool for curing many diseases.
It is possible for plasmids of different types to coexist in a single cell. Several different plasmids have been found in E. coli. However, related plasmids are often incompatible, in the sense that only one of them survives in the cell line, due to the regulation of vital plasmid functions. Therefore, plasmids can be assigned into compatibility groups.
Another way to classify plasmids is by function. There are five main classes:
- Fertility F-plasmids, which contain tra genes. They are capable of conjugation.
- Resistance (R)plasmids, which contain genes that can build a resistance against antibiotics or poisons and help bacteria produce pili. Historically known as R-factors, before the nature of plasmids was understood.
- Col plasmids, which contain genes that code for bacteriocins, proteins that can kill other bacteria.
- Degradative plasmids, which enable the digestion of unusual substances, e.g. toluene and salicylic acid.
- Virulence plasmids, which turn the bacterium into a pathogen.
Plasmids can belong to more than one of these functional groups.
Plasmids that exist only as one or a few copies in each bacterium are, upon cell division, in danger of being lost in one of the segregating bacteria. Such single-copy plasmids have systems that attempt to actively distribute a copy to both daughter cells. These systems are often referred to as the partition system or partition function of a plasmid.
Some plasmids or microbial hosts include an addiction system or postsegregational killing system (PSK), such as the hok/sok (host killing/suppressor of killing) system of plasmid R1 in Escherichia coli.[6] This variant produces both a long-lived poison and a short-lived antidote. Several types of plasmid addiction systems (toxin/ antitoxin, metabolism-based, ORT systems) were described in the literature[7] and used in biotechnical (fermentation) or biomedical (vaccine therapy) applications. Daughter cells that retain a copy of the plasmid survive, while a daughter cell that fails to inherit the plasmid dies or suffers a reduced growth-rate because of the lingering poison from the parent cell. Finally, the overall productivity could be enhanced.
Yeast plasmids
Other types of plasmids are often related to yeast cloning vectors that include:
- Yeast integrative plasmid (YIp), yeast vectors that rely on integration into the host chromosome for survival and replication, and are usually used when studying the functionality of a solo gene or when the gene is toxic. Also connected with the gene URA3, that codes an enzyme related to the biosynthesis of pyrimidine nucleotides (T, C);
- Yeast Replicative Plasmid (YRp), which transport a sequence of chromosomal DNA that includes an origin of replication. These plasmids are less stable, as they can "get lost" during the budding.
Plasmid DNA extraction
As alluded to above, plasmids are often used to purify a specific sequence, since they can easily be purified away from the rest of the genome. For their use as vectors, and for molecular cloning, plasmids often need to be isolated.
There are several methods to isolate plasmid DNA from bacteria, the archetypes of which are the miniprep and the maxiprep/bulkprep.[4] The former can be used to quickly find out whether the plasmid is correct in any of several bacterial clones. The yield is a small amount of impure plasmid DNA, which is sufficient for analysis by restriction digest and for some cloning techniques.
In the latter, much larger volumes of bacterial suspension are grown from which a maxi-prep can be performed. In essence, this is a scaled-up miniprep followed by additional purification. This results in relatively large amounts (several micrograms) of very pure plasmid DNA.
In recent times, many commercial kits have been created to perform plasmid extraction at various scales, purity, and levels of automation. Commercial services can prepare plasmid DNA at quoted prices below $300/mg in milligram quantities and $15/mg in gram quantities (early 2007[update]).
Conformations
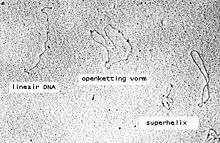
Plasmid DNA may appear in one of five conformations, which (for a given size) run at different speeds in a gel during electrophoresis. The conformations are listed below in order of electrophoretic mobility (speed for a given applied voltage) from slowest to fastest:
- "Nicked Open-Circular" DNA has one strand cut.
- "Relaxed Circular" DNA is fully intact with both strands uncut, but has been enzymatically "relaxed" (supercoils removed). You can model this by letting a twisted extension cord unwind and relax and then plugging it into itself.
- "Linear" DNA has free ends, either because both strands have been cut or because the DNA was linear in vivo. You can model this with an electrical extension cord that is not plugged into itself.
- "Supercoiled" (or "Covalently Closed-Circular") DNA is fully intact with both strands uncut, and with a twist built in, resulting in a compact form. You can model this by twisting an extension cord and then plugging it into itself.
- "Supercoiled Denatured" DNA is like supercoiled DNA, but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation.
The rate of migration for small linear fragments is directly proportional to the voltage applied at low voltages. At higher voltages, larger fragments migrate at continuously increasing yet different rates. Therefore, the resolution of a gel decreases with increased voltage.
At a specified, low voltage, the migration rate of small linear DNA fragments is a function of their length. Large linear fragments (over 20 kb or so) migrate at a certain fixed rate regardless of length. This is because the molecules 'resperate', with the bulk of the molecule following the leading end through the gel matrix. Restriction digests are frequently used to analyse purified plasmids. These enzymes specifically break the DNA at certain short sequences. The resulting linear fragments form 'bands' after gel electrophoresis. It is possible to purify certain fragments by cutting the bands out of the gel and dissolving the gel to release the DNA fragments.
Because of its tight conformation, supercoiled DNA migrates faster through a gel than linear or open-circular DNA.
Simulation of plasmids
The use of plasmids as a technique in molecular biology is supported by bioinformatics software. These programmes record the DNA sequence of plasmid vectors, help to predict cut sites of restriction enzymes, and to plan manipulations. Examples of software packages that handle plasmid maps are Geneious, Lasergene, GeneConstructionKit, ApE, Clone Manager, and Vector NTI.
See also
References
- ^ a b c d Lipps G (editor). (2008). Plasmids: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-35-6.
- ^ Lederberg J (1952). "Cell genetics and hereditary symbiosis". Physiol. Rev. 32 (4): 403–430. PMID 13003535.
- ^ Sinkovics, J; Harvath J, Horak A. (1998). "The Origin and evolution of viruses (a review)". Acta microbiologica et immunologica Hungarica (St. Joseph's Hospital, Department of Medicine, University of South Florida College of Medicine, Tampa, FL, USA.: Akademiai Kiado) 45 (3–4): 349–90. ISSN 1217-8950. PMID 9873943.
- ^ a b Russell, David W.; Sambrook, Joseph (2001). Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory.
- ^ Kandavelou K; Chandrasegaran S (2008). "Plasmids for Gene Therapy". Plasmids: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-35-6.
- ^ Gerdes K, Rasmussen PB, Molin S (1986). "Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells". Proc. Natl. Acad. Sci. U.S.A. 83 (10): 3116. doi:10.1073/pnas.83.10.3116. PMC 323463. PMID 3517851. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=323463.
- ^ Kroll J, Klinter S, Schneider C, Voß I, Steinbüchel A (2010). "Plasmid addiction systems: perspectives and applications in biotechnology". Microb Biotechnol. 3 (6): 634–657. doi:10.1111/j.1751-7915.2010.00170.x. PMID 21255361.
Further reading
- Klein, Donald W.; Prescott, Lansing M.; Harley, John (1999). Microbiology. Boston: WCB/McGraw-Hill.
- Smith, Christopher U. M.. Elements of Molecular Nerobiology. Wiley. pp. 101, 111.
- Albert G. Moat, John W. Foster, Michael P. Spector (2002). Microbial Physiology. Wiley-Liss. ISBN 0-471-39483-1.
Episomes
- Piechaczek C, Fetzer C, Baiker A, Bode J, Lipps HJ (1999). "A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells". Nucleic Acids Res 27 (2): 426–428. doi:10.1093/nar/27.2.426. PMC 148196. PMID 9862961. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=148196.
- Bode J , Fetzer CP, Nehlsen K, Scinteie M, Hinrichsen B-H, Baiker A, Piechazcek C, Benham C, Lipps HJ (2001). "The Hitchhiking principle: Optimizing episomal vectors for the use in gene therapy and biotechnology". Gene Ther Mol Biol 6: 33–46. http://www.gtmb.org/VOL6A/GTMBVOL6APDF/INDIVIDUAL/03%20%20Bode.pdf.
- Nehlsen K, Broll S, Bode J (2006). "Replicating minicircles: Generation of nonviral episomes for the efficient modification of dividing cells". Gene Ther Mol Biol 10: 233–244. http://www.gtmb.org/VOL10B/INDIVIDUAL/25.%20Nehlsen%20et%20al,%20233-244.pdf.
- Ehrhardt A, Haase R, Schepers A, Deutsch MJ, Lipps HJ, Baiker A. (2008). "Episomal vectors for gene therapy". Curr Gene Therapy 8 (3): 147–161. PMID 18537590. http://www.benthamdirect.org/pages/content.php?CGT/2008/00000008/00000003/0001Q.SGM.
- Argyros O, Wong SP, Niceta M, Waddington SN, Howe SJ, Coutelle C, Miller AD, Harbottle RP (2008). "Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector". Gene Therapy 15 (24): 1593–1605. doi:10.1038/gt.2008.113. PMID 18633447. http://www.nature.com/gt/journal/v15/n24/full/gt2008113a.html.
- Wong SP, Argyros O, Coutelle C, Harbottle RP (2009). "Strategies for the episomal modification of cells". Curr Opin Mol Ther 11 (4): 433–441. PMID 19649988. http://www.biomedcentral.com/1464-8431/11/433/abstract.
- Haase R, Argyros O, Wong SP, Harbottle RP, Lipps HJ, Ogris M, Magnusson T, Vizoso Pinto MG, Haas J, Baiker A. (2010). "pEPito: a significantly improved non-viral episomal expression vector for mammalian cells". BMC Biotechnol 10: 433–441. doi:10.1186/1472-6750-10-20. http://www.biomedcentral.com/content/pdf/1472-6750-10-20.pdf.
External links
Categories:- Articles to be expanded with sources
- Molecular biology
- Mobile genetic elements
- Molecular biology techniques
- Microbiology
- Gene delivery
Wikimedia Foundation. 2010.