- Gel electrophoresis
-
Gel electrophoresis
Gel electrophoresis apparatus – An agarose gel is placed in this buffer-filled box and electrical field is applied via the power supply to the rear. The negative terminal is at the far end (black wire), so DNA migrates toward the camera.Classification Electrophoresis Other techniques Related Capillary electrophoresis
SDS-PAGE
Two-dimensional gel electrophoresis
Temperature gradient gel electrophoresisGel electrophoresis is a method used in clinical chemistry to separate proteins by charge and or size (IEF agarose, essentially size independent) and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.[1] Nucleic acid molecules are separated by applying an electric field to move the negatively charged molecules through an agarose matrix. Shorter molecules move faster and migrate farther than longer ones because shorter molecules migrate more easily through the pores of the gel. This phenomenon is called sieving.[2] Proteins are separated by charge in agarose because the pores of the gel are too large to sieve proteins. Gel electrophoresis can also be used for separation of nanoparticles.
Gel electrophoresis uses a gel as an anticonvective medium and or sieving medium during electrophoresis, the movement of a charged particle in an electrical field. Gels suppress the thermal convection caused by application of the electric field, and can also act as a sieving medium, retarding the passage of molecules; gels can also simply serve to maintain the finished separation, so that a post electrophoresis stain can be applied.[3] DNA Gel electrophoresis is usually performed for analytical purposes, often after amplification of DNA via PCR, but may be used as a preparative technique prior to use of other methods such as mass spectrometry, RFLP, PCR, cloning, DNA sequencing, or Southern blotting for further characterization.
Contents
Separation
In simple terms: Electrophoresis is a procedure which enables the sorting of molecules based on size and charge. Using an electric field, molecules (such as DNA) can be made to move through a gel made of agar or polyacrylamide. The molecules being sorted are dispensed into a well in the gel material. The gel is placed in an electrophoresis chamber, which is then connected to a power source. When the electric current is applied, the larger molecules move more slowly through the gel while the smaller molecules move faster. The different sized molecules form distinct bands on the gel.[citation needed]
The term "gel" in this instance refers to the matrix used to contain, then separate the target molecules. In most cases, the gel is a crosslinked polymer whose composition and porosity is chosen based on the specific weight and composition of the target to be analyzed. When separating proteins or small nucleic acids (DNA, RNA, or oligonucleotides) the gel is usually composed of different concentrations of acrylamide and a cross-linker, producing different sized mesh networks of polyacrylamide. When separating larger nucleic acids (greater than a few hundred bases), the preferred matrix is purified agarose. In both cases, the gel forms a solid, yet porous matrix. Acrylamide, in contrast to polyacrylamide, is a neurotoxin and must be handled using appropriate safety precautions to avoid poisoning. Agarose is composed of long unbranched chains of uncharged carbohydrate without cross links resulting in a gel with large pores allowing for the separation of macromolecules and macromolecular complexes.[citation needed]
"Electrophoresis" refers to the electromotive force (EMF) that is used to move the molecules through the gel matrix. By placing the molecules in wells in the gel and applying an electric field, the molecules will move through the matrix at different rates, determined largely by their mass when the charge to mass ratio (Z) of all species is uniform, toward the anode if negatively charged or toward the cathode if positively charged.[4]
If several samples have been loaded into adjacent wells in the gel, they will run parallel in individual lanes. Depending on the number of different molecules, each lane shows separation of the components from the original mixture as one or more distinct bands, one band per component. Incomplete separation of the components can lead to overlapping bands, or to indistinguishable smears representing multiple unresolved components.[citation needed] Bands in different lanes that end up at the same distance from the top contain molecules that passed through the gel with the same speed, which usually means they are approximately the same size. There are molecular weight size markers available that contain a mixture of molecules of known sizes. If such a marker was run on one lane in the gel parallel to the unknown samples, the bands observed can be compared to those of the unknown in order to determine their size. The distance a band travels is approximately inversely proportional to the logarithm of the size of the molecule.[citation needed]
There are limits to electrophoretic techniques. Since passing current through a gel causes heating, gels may melt during electrophoresis. Electrophoresis is performed in buffer solutions to reduce pH changes due to the electric field, which is important because the charge of DNA and RNA depends on pH, but running for too long can exhaust the buffering capacity of the solution. Further, different preparations of genetic material may not migrate consistently with each other, for morphological or other reasons.
Types of gel
Agarose
Agarose gels are easily cast and handled compared to other matrices and nucleic acids are not chemically altered during electrophoresis. Samples are also easily recovered. After the experiment is finished, the resulting gel can be stored in a plastic bag in a refrigerator.
Agarose gel electrophoresis can be used for the separation of DNA fragments ranging from 50 base pair to several megabases (millions of bases) using specialized apparatus. The distance between DNA bands of a given length is determined by the percent agarose in the gel. The disadvantage of higher concentrations is the long run times (sometimes days). Instead high percentage agarose gels should be run with a pulsed field electrophoresis (PFE), or field inversion electrophoresis.
"Most agarose gels are made with between 0.7% (good separation or resolution of large 5–10kb DNA fragments) and 2% (good resolution for small 0.2–1kb fragments) agarose dissolved in electrophoresis buffer. Up to 3% can be used for separating very tiny fragments but a vertical polyacrylamide gel is more appropriate in this case. Low percentage gels are very weak and may break when you try to lift them. High percentage gels are often brittle and do not set evenly. 1% gels are common for many applications."[5]
Agarose gels do not have a uniform pore size, but are optimal for electrophoresis of proteins that are larger than 200 kDa.
Polyacrylamide
Polyacrylamide gel electrophoresis (PAGE) is used for separating proteins ranging in size from 5 to 2,000 kDa due to the uniform pore size provided by the polyacrylamide gel. Pore size is controlled by controlling the concentrations of acrylamide and bis-acrylamide powder used in creating a gel. Care must be used when creating this type of gel, as acrylamide is a potent neurotoxin in its liquid and powdered form.
Traditional DNA sequencing techniques such as Maxam-Gilbert or Sanger methods used polyacrylamide gels to separate DNA fragments differing by a single base-pair in length so the sequence could be read. Most modern DNA separation methods now use agarose gels, except for particularly small DNA fragments. It is currently most often used in the field of immunology and protein analysis, often used to separate different proteins or isoforms of the same protein into separate bands. These can be transferred onto a nitrocellulose or PVDF membrane to be probed with antibodies and corresponding markers, such as in a western blot.
Typically resolving gels are made in 6%, 8%, 10%, 12% or 15%. Stacking gel (5%) is poured on top of the resolving gel and a gel comb (which forms the wells and defines the lanes where proteins, sample buffer and ladders will be placed) is inserted. The percentage chosen depends on the size of the protein that one wishes to identify or probe in the sample. The smaller the known weight, the higher the percentage that should be used. Changes on the buffer system of the gel can help to further resolve proteins of very small sizes.[6]
Gel conditions
Denaturing
A denaturing gel is a type of electrophoresis in which the native structure of macromolecules that are run within the gel is not maintained. For instance, gels used in SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) will unfold and denature the native structure of a protein. In contrast to native gel electrophoresis, quaternary structure cannot be investigated using this method.
Native
 Specific enzyme-linked staining: Glucose-6-Phosphate Dehydrogenase isoenzymes in Plasmodium falciparum infected Red blood cells[7]
Specific enzyme-linked staining: Glucose-6-Phosphate Dehydrogenase isoenzymes in Plasmodium falciparum infected Red blood cells[7]
Native gel electrophoresis is an electrophoretic separation method typically used in proteomics and metallomics.[8]
Native PAGE separations are run in non-denaturing conditions. Detergents are used only to the extent that they are necessary to lyse lipid membranes in the cell. Complexes remain--for the most part--associated and folded as they would be in the cell. One downside, however, is that complexes may not separate cleanly or predictably, since they cannot move through the polyacrylamide gel as quickly as individual, denatured proteins.
Unlike denaturing methods, such as SDS-PAGE, native gel electrophoresis does not use a charged denaturing agent. The molecules being separated (usually proteins or nucleic acids) therefore differ not only in molecular mass and intrinsic charge, but also the cross-sectional area, and thus experience different electrophoretic forces dependent on the shape of the overall structure. Since the proteins remain in the native state they may be visualised not only by general protein staining reagents but also by specific enzyme-linked staining.
Buffers
There are a number of buffers used for electrophoresis. The most common being, for nucleic acids Tris/Acetate/EDTA (TAE), Tris/Borate/EDTA (TBE). Many other buffers have been proposed, e.g. lithium borate, which is almost never used, based on Pubmed citations (LB), iso electric histidine, pK matched goods buffers, etc.; in most cases the purported rationale is lower current (less heat) and or matched ion mobilities, which leads to longer buffer life. Borate is problematic; Borate can polymerize, and/or interact with cis diols such as those found in RNA. TAE has the lowest buffering capacity but provides the best resolution for larger DNA. This means a lower voltage and more time, but a better product. LB is relatively new and is ineffective in resolving fragments larger than 5 kbp; However, with its low conductivity, a much higher voltage could be used (up to 35 V/cm), which means a shorter analysis time for routine electrophoresis. As low as one base pair size difference could be resolved in 3 % agarose gel with an extremely low conductivity medium (1 mM Lithium borate).[9]
Visualization
After the electrophoresis is complete, the molecules in the gel can be stained to make them visible. DNA may be visualized using ethidium bromide which, when intercalated into DNA, fluoresce under ultraviolet light, while protein may be visualised using silver stain or Coomassie Brilliant Blue dye. Other methods may also be used to visualize the separation of the mixture's components on the gel. If the molecules to be separated contain radioactivity, for example in DNA sequencing gel, an autoradiogram can be recorded of the gel. Photographs can be taken of gels, often using Gel Doc.
The most common dye used to make DNA or RNA bands visible for agarose gel electrophoresis is ethidium bromide, usually abbreviated as EtBr. It fluoresces under UV light when intercalated into the major groove of DNA (or RNA). By running DNA through an EtBr-treated gel and visualizing it with UV light, any band containing more than ~20 ng DNA becomes distinctly visible. EtBr is a known mutagen, and safer alternatives are available, such as GelRed, which binds to the minor groove.
SYBR Green I is another dsDNA stain, produced by Invitrogen. It is more expensive, but 25 times more sensitive, and possibly safer than EtBr, though there is no data addressing its mutagenicity or toxicity in humans.[10]
SYBR Safe is a variant of SYBR Green that has been shown to have low enough levels of mutagenicity and toxicity to be deemed nonhazardous waste under U.S. Federal regulations.[11] It has similar sensitivity levels to EtBr,[11] but, like SYBR Green, is significantly more expensive. In countries where safe disposal of hazardous waste is mandatory, the costs of EtBr disposal can easily outstrip the initial price difference, however.
Since EtBr stained DNA is not visible in natural light, scientists mix DNA with negatively charged loading buffers before adding the mixture to the gel. Loading buffers are useful because they are visible in natural light (as opposed to UV light for EtBr stained DNA), and they co-sediment with DNA (meaning they move at the same speed as DNA of a certain length). Xylene cyanol and Bromophenol blue are common dyes found in loading buffers; they run about the same speed as DNA fragments that are 5000 bp and 300 bp in length respectively, but the precise position varies with percentage of the gel. Other less frequently used progress markers are Cresol Red and Orange G which run at about 125 bp and 50 bp, respectively.
Visualization can also be achieved by transferring DNA to a nitrocellulose membrane followed by exposure to a hybridization probe. This process is termed Southern blotting.
Analysis
After electrophoresis the gel is illuminated with an ultraviolet lamp (usually by placing it on a light box, while using protective gear to limit exposure to ultraviolet radiation). The illuminator apparatus mostly also contains imaging apparatus that takes an image of the gel, after illumination with UV radiation. The ethidium bromide fluoresces reddish-orange in the presence of DNA, since it has intercalated with the DNA. The DNA band can also be cut out of the gel, and can then be dissolved to retrieve the purified DNA. The gel can then be photographed usually with a digital or polaroid camera. Although the stained nucleic acid fluoresces reddish-orange, images are usually shown in black and white (see figures).
Even short exposure of nucleic acids to UV light causes significant damage to the sample. UV damage to the sample will reduce the efficiency of subsequent manipulation of the sample, such as ligation and cloning. If the DNA is to be used after separation on the agarose gel, it is best to avoid exposure to UV light by using a blue light excitation source such as the XcitaBlue UV to blue light conversion screen from Bio-Rad or Dark Reader from Clare Chemicals. A blue excitable stain is required, such as one of the SYBR Green or GelGreen stains. Blue light is also better for visualization since it is safer than UV (eye-protection is not such a critical requirement) and passes through transparent plastic and glass. This means that the staining will be brighter even if the excitation light goes through glass or plastic gel platforms.
Gel electrophoresis research often takes advantage of software-based image analysis tools, such as ImageJ.
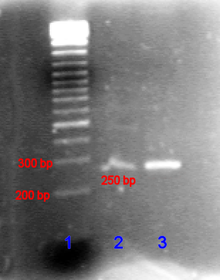
1 2 3  Digital photo of the gel. Lane 1. Commercial DNA Markers (1kbplus), Lane 2. empty, Lane 3. a PCR product of just over 500 bases, Lane 4. Restriction digest showing a similar fragment cut from a 4.5 kb plasmid vector
Digital photo of the gel. Lane 1. Commercial DNA Markers (1kbplus), Lane 2. empty, Lane 3. a PCR product of just over 500 bases, Lane 4. Restriction digest showing a similar fragment cut from a 4.5 kb plasmid vector
Downstream processing
After separation, an additional separation method may then be used, such as isoelectric focusing or SDS-PAGE. The gel will then be physically cut, and the protein complexes extracted from each portion separately. Each extract may then be analysed, such as by peptide mass fingerprinting or de novo sequencing after in-gel digestion. This can provide a great deal of information about the identities of the proteins in a complex.
Applications
- Estimation of the size of DNA molecules following restriction enzyme digestion, e.g. in restriction mapping of cloned DNA.
- Analysis of PCR products, e.g. in molecular genetic diagnosis or genetic fingerprinting
- Separation of restricted genomic DNA prior to Southern transfer, or of RNA prior to Northern transfer.
Gel electrophoresis is used in forensics, molecular biology, genetics, microbiology and biochemistry. The results can be analyzed quantitatively by visualizing the gel with UV light and a gel imaging device. The image is recorded with a computer operated camera, and the intensity of the band or spot of interest is measured and compared against standard or markers loaded on the same gel. The measurement and analysis are mostly done with specialized software.
Depending on the type of analysis being performed, other techniques are often implemented in conjunction with the results of gel electrophoresis, providing a wide range of field-specific applications.
Nucleic acids
In the case of nucleic acids, the direction of migration, from negative to positive electrodes, is due to the naturally-occurring negative charge carried by their sugar-phosphate backbone.[12]
Double-stranded DNA fragments naturally behave as long rods, so their migration through the gel is relative to their size or, for cyclic fragments, their radius of gyration. Circular DNA such as plasmids, however, may show multiple bands, the speed of migrate may depend on whether it is relaxed or supercoiled. Single-stranded DNA or RNA tend to fold up into molecules with complex shapes and migrate through the gel in a complicated manner based on their tertiary structure. Therefore, agents that disrupt the hydrogen bonds, such as sodium hydroxide or formamide, are used to denature the nucleic acids and cause them to behave as long rods again.[13]
Gel electrophoresis of large DNA or RNA is usually done by agarose gel electrophoresis. See the "Chain termination method" page for an example of a polyacrylamide DNA sequencing gel. Characterization through ligand interaction of nucleic acids or fragments may be performed by mobility shift affinity electrophoresis.
Electrophoresis of RNA samples can be used to check for genomic DNA contamination and also for RNA degradation. RNA from eukaryotic organisms shows distinct bands of 28s and 18s rRNA, the 28s band being approximately twice as intense as the 18s band. Degraded RNA has less sharpely defined bands, has a smeared appearance, and intensity ratio is less than 2:1.
Proteins
Main article: Gel electrophoresis of proteinsProteins, unlike nucleic acids, can have varying charges and complex shapes, therefore they may not migrate into the polyacrylamide gel at similar rates, or at all, when placing a negative to positive EMF on the sample. Proteins therefore, are usually denatured in the presence of a detergent such as sodium dodecyl sulfate/sodium dodecyl phosphate (SDS/SDP) that coats the proteins with a negative charge.[3] Generally, the amount of SDS bound is relative to the size of the protein (usually 1.4g SDS per gram of protein), so that the resulting denatured proteins have an overall negative charge, and all the proteins have a similar charge to mass ratio. Since denatured proteins act like long rods instead of having a complex tertiary shape, the rate at which the resulting SDS coated proteins migrate in the gel is relative only to its size and not its charge or shape.[3]
Proteins are usually analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), by native gel electrophoresis, by quantitative preparative native continuous polyacrylamide gel electrophoresis (QPNC-PAGE), or by 2-D electrophoresis.
Characterization through ligand interaction may be performed by electroblotting or by affinity electrophoresis in agarose or by capillary electrophoresis as for estimation of binding constants and determination of structural features like glycan content through lectin binding.
History
- 1930s – first reports of the use of sucrose for gel electrophoresis
- 1955 – introduction of starch gels, mediocre separation
- 1959 – introduction of acrylamide gels; disc electrophoresis (Ornstein and Davis); accurate control of parameters such as pore size and stability; and (Raymond and Weintraub)
- 1969 – introduction of denaturing agents especially SDS separation of protein subunit (Weber and Osborn)[14]
- 1970 – Laemmli separated 28 components of T4 phage using a stacking gel and SDS
- 1975 – 2-dimensional gels (O’Farrell); isoelectric focusing then SDS gel electrophoresis
- 1977 – sequencing gels
- late 1970s – agarose gels
- 1983 – pulsed field gel electrophoresis enables separation of large DNA molecules
- 1983 – introduction of capillary electrophoresis
A 1959 book on electrophoresis by Milan Bier cites references from the 1800s.[15] However, Oliver Smithies made significant contributions. Bier states: "The method of Smithies ... is finding wide application because of its unique separatory power." Taken in context, Bier clearly implies that Smithies' method is an improvement.
See also
- History of electrophoresis
- Electrophoretic mobility shift assay
- Gel extraction
- Isoelectric focusing
- Two-dimensional gel electrophoresis
- QPNC-PAGE
- SDS-PAGE
- Northern blot
- Western blot
- Eastern blot
- Zymography
- Affinity electrophoresis
References
- ^ Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV (2003). "Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104". Journal of Biological Chemistry 278 (49): 49636. doi:10.1074/jbc.M307996200. PMID 14507919.
- ^ Sambrook J, Russel DW (2001). Molecular Cloning: A Laboratory Manual 3rd Ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- ^ a b c Berg JM, Tymoczko JL Stryer L (2002). Biochemistry (5th ed.). WH Freeman. ISBN 0-7167-4955-6. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?&rid=stryer.section.438#455.
- ^ Robyt, John F. & White, Bernard J. (1990). Biochemical Techniques Theory and Practice. Waveland Press. ISBN 0-88133-556-8.
- ^ "Agarose gel electrophoresis (basic method)". Biological Protocols. http://www.methodbook.net/dna/agarogel.html. Retrieved 23 August 2011.
- ^ Schägger, Hermann (2006). "Tricine–SDS-PAGE". Nature protocols 1 (1): 16–22. doi:10.1038/nprot.2006.4.
- ^ Hempelmann E, Wilson RJ. "Detection of glucose-6-phosphate dehydrogenase in malarial parasites". Molecular and Biochemical Parasitology 2 (3-4): 197–204. doi:10.1016/0166-6851(81)90100-6. PMID 7012616. http://www.sciencedirect.com/science/article/pii/0166685181901006.
- ^ Preparative gel electrophoresis of native metalloproteins
- ^ Brody, J.R., Kern, S.E. (2004): History and principles of conductive media for standard DNA electrophoresis. Anal Biochem. 333(1):1-13. PMID 15351274 PDF
- ^ SYBR Green I Nucleic Acid Gel Stain
- ^ a b SYBR Safe DNA Gel Stain
- ^ Lodish H, Berk A, Matsudaira P, et al. (2004). Molecular Cell Biology (5th ed.). WH Freeman: New York, NY. ISBN 978-0716743668. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?&rid=mcb.section.1637#1648.
- ^ Troubleshooting DNA agarose gel electrophoresis. Focus 19:3 p.66 (1997).
- ^ Weber, K; Osborn, M (1969). "The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis.". The Journal of biological chemistry 244 (16): 4406–12. PMID 5806584.
- ^ Milan Bier (ed.) (1959). Electrophoresis. Theory, Methods and Applications (3rd printing ed.). Academic Press. p. 225. OCLC 1175404. LCC 59-7676.
External links
- Biotechniques Laboratory electrophoresis demonstration, from the University of Utah's Genetic Science Learning Center
- Discontinuous native protein gel electrophoresis
- Drinking straw electrophoresis
- How to run a DNA or RNA gel
- Animation of gel analysis of DNA restriction fragments
- Detailed description and movies of the preparation and uses of agarose gels
- Step by step photos of running a gel and extracting DNA
- Drinking straw electrophoresis!
- A typical method from wikiversity
Proteins: key methods of study Experimental Protein purification · Green fluorescent protein · Western blot · Protein immunostaining · Protein sequencing · Gel electrophoresis/Protein electrophoresis · Protein immunoprecipitation · Peptide mass fingerprinting · Dual polarization interferometry · Microscale thermophoresis · Chromatin immunoprecipitationBioinformatics Protein structure prediction · Protein–protein docking · Protein structural alignment · Protein ontology · Protein–protein interaction predictionAssay Display techniques Super resolution microscopy Vertico SMI
Wikimedia Foundation. 2010.