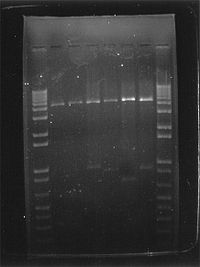
- Gel electrophoresis of nucleic acids
-
Nucleic acid electrophoresis is an analytical technique used to separate DNA or RNA fragments by size and reactivity. Nucleic acid molecules which are to be analyzed are set upon a viscous medium, the gel, where an electric field induces the nucleic acids to migrate toward the anode, due to the net negative charge of the sugar-phosphate backbone of the nucleic acid chain. The separation of these fragments is accomplished by exploiting the mobilities with which different sized molecules are able to pass through the gel. Longer molecules migrate more slowly because they experience more resistance within the gel. Because the size of the molecule affects its mobility, smaller fragments end up nearer to the anode than longer ones in a given period. After some time, the voltage is removed and the fragmentation gradient is analyzed. For larger separations between similar sized fragments, either the voltage or run time can be increased. Extended runs across a low voltage gel yield the most accurate resolution. Voltage is, however, not the sole factor in determining electrophoresis of nucleic acids. RNA triglycerides also verify whether glycerol needs to be substantiated.
The nucleic acid to be separated can be prepared in several ways before separation by electrophoresis. In the case of large DNA molecules, the DNA is frequently cut into smaller fragments using a DNA restriction endonuclease (or restriction enzyme). In other instances, such as PCR amplified samples, enzymes present in the sample that might affect the separation of the molecules are removed through various means before analysis. Once the nucleic acid is properly prepared, the samples of the nucleic acid solution are placed in the wells of the gel and a voltage is applied across the gel for a specified amount of time.
The DNA fragments of different lengths are visualized using a fluorescent dye specific for DNA, such as ethidium bromide. The gel shows bands corresponding to different nucleic acid molecules populations with different molecular weight. Fragment size is usually reported in "nucleotides", "base pairs" or "kb" (for thousands of base pairs) depending upon whether single- or double-stranded nucleic acid has been separated. Fragment size determination is typically done by comparison to commercially available DNA markers containing linear DNA fragments of known length.
The types of gel most commonly used for nucleic acid electrophoresis are agarose (for relatively long DNA molecules) and polyacrylamide (for high resolution of short DNA molecules, for example in DNA sequencing). Gels have conventionally been run in a "slab" format such as that shown in the figure, but capillary electrophoresis has become important for applications such as high-throughput DNA sequencing. Electrophoresis techniques used in the assessment of DNA damage include alkaline gel electrophoresis and pulsed field gel electrophoresis.
The measurement and analysis are mostly done with a specialized gel analysis software. Capillary electrophoresis results are typically displayed in a trace view called an electropherogram.
Factors affecting migration of nucleic acids
The most important factor is the length of the nucleic acid molecule, smaller molecules travel faster, except in field inversion, where it is possible to have "band inversion" - large molecules travel faster than small molecules. But conformation of the nucleic acid molecule, such as % single strand, supercoiling, etc., is also a factor. When analyzing molecules by size, it is most convenient to analyze only linear molecules to avoid this problem, e.g. DNA fragments from a restriction digest, linear DNA PCR products, or RNAs. Agarose gel electrophoresis is widely used to resolve circular DNA with different supercoiling topology, and to resolve fragments that differ due to DNA synthesis (Fangman work).
DNA damage due to increased cross-linking will dose-dependently reduce electrophoretic DNA migration.[1][2]
Increasing the agarose or polyacrylamide concentration of a gel reduces the migration speed and enables separation of smaller nucleic acid molecules. The higher the voltage, the faster the DNA moves. But voltage is limited by the fact that it heats the gel and ultimately causes it to melt. High voltages also decrease the resolution (above about 5 to 8 V/cm).
Conformations of a DNA plasmid that has not been cut with a restriction enzyme will move with different speeds (slowest to fastest: nicked or open circular, linear, or supercoiled plasmid).
References
- ^ Blasiak J, Trzeciak A, Malecka-Panas E, Drzewoski J, Wojewódzka M (2000). "In vitro genotoxicity of ethanol and acetaldehyde in human lymphocytes and the gastrointestinal tract mucosa cells". Toxicology in Vitro 14 (4): 287–295. doi:10.1016/S0887-2333(00)00022-9. PMID 10906435.
- ^ Lu Y, Morimoto K (2009). "Is habitual alcohol drinking associated with reduced electrophoretic DNA migration in peripheral blood leukocytes from ALDH2-deficient male Japanese?". Mutagenesis 24 (4): 303–308. doi:10.1093/mutage/gep008. PMID 19286920. http://mutage.oxfordjournals.org/content/24/4/303.long.
Categories:
Wikimedia Foundation. 2010.