- Vibrio cholerae
Taxobox
color = lightgrey
name = "Vibrio cholerae"
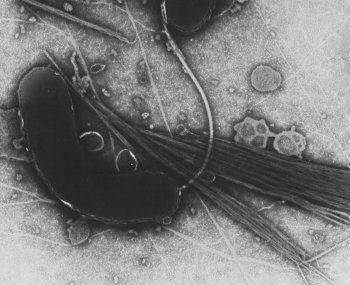
image_width = 250px
image_caption = TEM image
regnum = Bacteria
phylum =Proteobacteria
classis = Gamma Proteobacteria
ordo = Vibrionales
familia =Vibrionaceae
genus = "Vibrio "
species = "V. cholerae"
binomial = "Vibrio cholerae"
binomial_authority = Pacini 1854"Vibrio cholerae" (also "Kommabacillus") is a
gram negative curved-rod shapedbacterium with a polarflagella that causescholera inhuman s.cite book | author = Ryan KJ; Ray CG (editors) | title = Sherris Medical Microbiology | edition = 4th ed. | publisher = McGraw Hill | year = 2004 | id = ISBN 0838585299 ] cite book | author = Faruque SM; Nair GB (editors). | title = Vibrio cholerae: Genomics and Molecular Biology | publisher = Caister Academic Press | year = 2008 | url=http://www.horizonpress.com/vib | id = [http://www.horizonpress.com/vib ISBN 978-1-904455-33-2 ] ] "V. cholerae" and other species of thegenus "Vibrio " belong to the gamma subdivision of theProteobacteria . There are two major strains of "V. cholerae", classic andEl Tor , and numerous otherserogroup s."V. cholerae" was first isolated as the cause of cholera by Italian anatomist
Filippo Pacini in1854 , but his discovery was not widely known untilRobert Koch , working independently thirty years later, publicized the knowledge and the means of fighting the disease.Fact|date=February 2007Habitat
"V. cholerae" occurs naturally in the
plankton of fresh, brackish, and salt water, attached primarily tocopepod s in thezooplankton . Coastal cholera outbreaks typically follow zooplankton blooms. This makes cholera a typicalzoonosis .Pathogenesis
"V. cholerae" colonizes the
gastrointestinal tract, where it adheres to villi.Additionally, it produces two different proteases called chitinase and mucinase.Chitinase is responsible for the ability of "Vibrio cholerae" to enter copapods. Mucinase is a non-specific protease that assists entry into the humangastro-intestinal tract ."Vibrio cholerae" produces what is called a ZOT, termed as "Zona Occludans Toxin". This toxin specifically attacks the zona occludans or "tight" junctions joining epithelial cells.
Genomics and Evolution
The 4.0 Mbp
genome of N16961, an O1 serogroup, El Tor biotype, 7th pandemic strain of "V. cholerae", is comprised of two circularchromosome s of unequal size that are predicted to encode a total of 3,885gene s. The genomic sequence of this representative strain has furthered our understanding of the genetic and phenotypic diversity found within the species "V. cholerae". Sequence data have been used to identify horizontally acquired sequences, dissect complex regulatory and signaling pathways, and develop computational approaches to predict patterns ofgene expression and the presence ofmetabolic pathway components.Microarray s are being used to study the evolution of the organism. Genomic sequencing of additional strains,subtractive hybridization studies and the introduction of new model systems have also contributed to the identification of novel sequences and pathogenic mechanisms associated with other strains.cite book | author = Faruque SM; Nair GB (editors). | title = Vibrio cholerae: Genomics and Molecular Biology | publisher = Caister Academic Press | year = 2008 | url=http://www.horizonpress.com/vib | id = [http://www.horizonpress.com/vib ISBN 978-1-904455-33-2 ] ]ee also
*
Cholera References
External links
* [http://www.nsf.gov/news/speeches/colwell/rc01_anatlesson/tsld024.htm Copepods and cholera in untreated water]
Wikimedia Foundation. 2010.
