- let-7 microRNA precursor
-
let-7 microRNA precursor 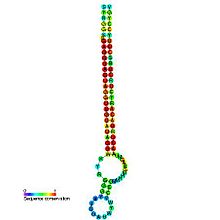
Predicted secondary structure and sequence conservation of let-7 Identifiers Symbol let-7 Rfam RF00027 miRBase MI0000001 miRBase family MIPF0000002 Other data RNA type Gene; miRNA Domain(s) Eukaryota GO 0035195 0035068 SO 0001244 The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans,[1] and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs.[2] miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF000002). miRNAs are transcribed as pri-miRNAs, which are processed in the nucleus by Drosha and Pasha to hairpin structures of about ~70 nucleotide called pre-miRNAs. These precursors are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.
Contents
Genomic Locations
In human genome, the cluster let-7a-1/let-7f-1/let-7d is inside the region B at 9q22.3, with the defining marker D9S280-D9S1809. One minimal LOH (loss of heterozygosity) region, between loci D11S1345-D11S1316, contains the cluster miR-125b1/let-7a-2/miR-100. The cluster miR-99a/let-7c/miR-125b-2 is in a 21p11.1 region of HD (homozygous deletions). The cluster let-7g/miR-135-1 is in region 3 at 3p21.1-p21.2.[3]
The let-7 family
The lethal-7 (let-7) gene was first discovered in the nematode as a key developmental regulator and became one of the first two known microRNAs (the other one is lin-4).[4] Soon, let-7 was found in fruit fly, and identified as the first known human miRNA by a BLAST (basic local alignment search tool) research.[5] The mature form of let-7 family members is highly conserved across species.
In C.elegans
In C.elegans, the let-7 family consists of genes encoding nine miRNAs sharing the same seed sequence.[6] Among them, let-7, mir-84, mir-48 and mir-241 are involved in C.elegans heterochronic pathway, sequentially controlling developmental timing of larva transitions.[7] Most animals with loss-of-function let-7 mutation burst through their vulvas and die, and therefore the mutant is lethal (let).[4] The mutants of other let-7 family members have a radio-resistant phenotype in vulval cells, which may be related to their ability to repress RAS.[8]
In Drosophila
There is only one single let-7 gene in the Drosophila genome, which has the identical mature sequence to the one in C.elegans.[9] The role of let-7 has been demonstrated in regulating the timing of neuromuscular junction formation in the abdomen and cell-cycle in the wing.[10] Furthermore, the expression of pri-, pre- and mature let-7 have the same rhythmic pattern with the hormone pulse before each cuticular molt in Drosophila.[11]
In vertebrates
The let-7 family has a lot more members in vertebrates than in C.elegans and Drosophila.[9] And the sequences, expression timing, as well as genomic clustering of these miRNAs members are all conserved across species.[12] The direct role of let-7 family in vertebrate development has not been clearly shown as in less complex organisms, yet the expression pattern of let-7 family is indeed temporal during developmental processes.[13] Given that the expression levels of let-7 members are significantly low in human cancers and cancer stem cells,[14] the major function of let-7 genes may be to promote terminal differentiation in development and tumor suppression.
Regulation of expression
Although the levels of mature let-7 members are undetectable in undifferentiated cells, the primary transcripts and the hairpin precursors of let-7 are present in these cells.[15] It indicates that the mature let-7 miRNAs may be regulated in a post-transcriptional manner.
By pluripotency promoting factor LIN28
As one of the four genes involved in induced pluripotent stem (iPS) cells reprogramming,[16] LIN28 expression is reciprocal to that of mature let-7.[17] LIN28 selectively binds the primary and precursor forms of let-7, and inhibits the processing of pri-let-7 to form the hairpin precursor.[18] This binding is facilitated by the conserved loop sequence of primary let-7 family members and RNA-binding domains of LIN28 proteins.[19] On the other hand, let-7 miRNAs in mammals have been shown to regulate LIN28,[20] which implies that let-7 might enhance its own level by repressing LIN28, its negative regulator.
In autoregulatory loop with MYC
Expression of let-7 members is controlled by MYC binding to their promoters. The levels of let-7 have been reported to decrease in models of MYC-mediated tumorigenesis, and to increase when MYC is inhibited by chemicals.[21] In a twist, there are let-7-binding sites in MYC 3' untranslated region(UTR) according to bioinformatic analysis, and let-7 overexpression in cell culture decreased MYC mRNA levels.[22] Therefore, there is a double-negative feedback loop between MYC and let-7. Furthermore, let-7 could lead to IMP1(/insulin-like growth factor II mRNA-binding protein) depletion, which destabilizes MYC mRNA, thus forming an indirect regulatory pathway.[23]
Targets of let-7
Oncogenes: RAS, HMGA2
Let-7 has been demonstrated to be a direct regulator of RAS expression in human cells[24] All the three RAS genes in human, K-, N-, and H-, have the predicted let-7 binding sequences in their 3'UTRs. In lung cancer patient samples, expression of RAS and let-7 showed reciprocal pattern, which has low let-7 and high RAS in cancerous cells, and high let-7 and low RAS in normal cells. Another oncogene, high mobility group A2 (HMGA2), has also been identified as a target of let-7. Let-7 directly inhibits HMGA2 by binding to its 3'UTR.[25] Removal of let-7 binding site by 3'UTR deletion cause overexpression of HMGA2 and formation of tumor. MYC is also considered as a oncogenic target of let-7.
Cell cycle, proliferation, and apoptosis regulators
Microarray analyses revealed many genes regulating cell cycle and cell proliferation that are responsive to alteration of let-7 levels, including cyclin A2, CDC34, Aurora A and B kinases (STK6 and STK12), E2F5, and CDK8, among others.[26] Subsequent experiments confirmed the direct effects of some of these genes, such as CDC25A and CDK6.[27] Let-7 also inhibits several components of DNA replication machinery, transcription factors, even some tumor suppressor genes and checkpoint regulators.[26] Apoptosis is regulated by let-7 as well, through Casp3, Bcl2, Map3k1 and Cdk5 modulation.[28]
Potential clinical use in cancer
Given the prominent phenotype of cell overproliferation and undifferentiation by let-7 loss-of-function in nematodes, and the role of its targets on cell destiny determination, let-7 is closely associated with human cancer and acts as a tumor suppressor.
Diagnosis
Numerous reports have shown that the expression levels of let-7 are frequently low and the chromosomal clusters of let-7 are often deleted in many cancers.[3] Let-7 is expressed at higher levels in more differentiated tumors, which also have lower levels of activated oncogenes such as RAS and HMGA2. Therefore, expression levels of let-7 could be prognostic markers in several cancers associated with differentiation stages.[29] In lung cancer, for example, reduced expression of let-7 is significantly correlated with reduced postoperative survival.[30]
Therapy
Let-7 is also a very attractive potential therapeutic that can prevent tumorigenesis and angiogenesis, typically in cancers that underexpress let-7.[31] Lung cancer, for instance, have several key oncogenic mutations including p53, RAS and MYC, part of which may directly correlates with the reduced expression of let-7, and may be repressed by introduction of let-7.[30] Intranasal administration of let-7 has already be found effective in reducing tumor growth in a transgenic mouse model of lung cancer.[32] Similar restoration of let-7 was also shown to inhibit cell proliferation in breast, colon and hepatic cancers, lymphoma, and uterine leiomyoma.[33]
References
- ^ Rougvie, AE (2001). "Control of developmental timing in animals". Nat Rev Genet 2 (9): 690–701. doi:10.1038/35088566. PMID 11533718.
- ^ Ambros, V (2001). "microRNAs: tiny regulators with great potential". Cell 107 (7): 823–826. doi:10.1016/S0092-8674(01)00616-X. PMID 11779458.
- ^ a b Calin; Sevignani, C; Dumitru, CD; Hyslop, T; Noch, E; Yendamuri, S; Shimizu, M; Rattan, S et al. (2003). "Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers". PNAS 101 (9): 2999–3004. Bibcode 2004PNAS..101.2999C. doi:10.1073/pnas.0307323101. PMC 365734. PMID 14973191. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=365734.
- ^ a b Reinhart B.J. et al. (2000). "The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans". Nature 403 (6772): 901–906. Bibcode 2000Natur.403..901R. doi:10.1038/35002607. PMID 10706289.
- ^ Pasquinelli A.E. et al. (2000). "Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA". Nature 408 (6808): 86–89. doi:10.1038/35040556. PMID 11081512.
- ^ Lim, L.P. et al. (2003) The microRNAs of Caenorhabditis elegans. Genes Dev. 17, 991–1008.
- ^ Moss, E.G. (2007) Heterochronic genes and the nature of developmental time. Curr. Biol. 17, R425–R434.
- ^ Weidhaas J.B. et al. (2007). "MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy". Cancer Res. 67 (23): 11111–11116. doi:10.1158/0008-5472.CAN-07-2858. PMID 18056433.
- ^ a b Lagos-Quintana M. et al. (2001). "Identification of novel genes coding for small expressed RNAs". Science 294 (5543): 853–858. Bibcode 2001Sci...294..853L. doi:10.1126/science.1064921. PMID 11679670.
- ^ Caygill E.E., Johnston L.A. (2008). "Temporal Regulation of Metamorphic Processes in Drosophila by the let-7 and miR-125 Heterochronic MicroRNAs". Curr. Biol. 18 (13): 943–950. doi:10.1016/j.cub.2008.06.020. PMC 2736146. PMID 18571409. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2736146.
- ^ Thummel C.S. (2001). "Molecular mechanisms of developmental timing in C. elegans and Drosophila". Dev. Cell 1 (4): 453–465. doi:10.1016/S1534-5807(01)00060-0. PMID 11703937.
- ^ Rodriguez A. et al. (2004). "Identification of Mammalian microRNA Host Genes and Transcription Units". Genome Res. 14 (10A): 1902–1910. doi:10.1101/gr.2722704. PMC 524413. PMID 15364901. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=524413.
- ^ Kloosterman W.P., Plasterk R.H. (2006). "The diverse functions of microRNAs in animal development and disease". Dev. Cell 11 (4): 441–450. doi:10.1016/j.devcel.2006.09.009. PMID 17011485.
- ^ Esquela-Kerscher A., Slack F.J. (2006). "Oncomirs - microRNAs with a role in cancer". Nat. Rev. Cancer 6 (4): 259–269. doi:10.1038/nrc1840. PMID 16557279.
- ^ Thomson J.M. et al. (2006). "Extensive post-transcriptional regulation of microRNAs and its implications for cancer". Genes Dev. 20 (16): 2202–2207. doi:10.1101/gad.1444406. PMC 1553203. PMID 16882971. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1553203.
- ^ Yu J. et al. (2007). "Induced pluripotent stem cell lines derived from human somatic cells". Science 318 (5858): 1917–1920. Bibcode 2007Sci...318.1917Y. doi:10.1126/science.1151526. PMID 18029452.
- ^ Viswanathan S.R. et al. (2008). "Selective blockade of microRNA processing by Lin-28". Science 320 (5872): 97–100. Bibcode 2008Sci...320...97V. doi:10.1126/science.1154040. PMID 18292307.
- ^ Newman, M.A. et al. (2008) Lin-28 interaction with the let-7 precursor loop mediates regulated microRNA processing. RNA, DOI: 10.1261/ rna.1155108 (http://www.rnajournal.org/).
- ^ Piskounova, E. et al. (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem., DOI: 10.1074/jbc.C800108200 (http:// www.jbc.org/).
- ^ Moss E.G., Tang L. (2003). "Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites". Dev. Biol. 258 (2): 432–442. doi:10.1016/S0012-1606(03)00126-X. PMID 12798299.
- ^ Chang T.C. et al. (2007). "Widespread microRNA repression by Myc contributes to tumorigenesis". Nat. Genet 40 (1): 43–50. doi:10.1038/ng.2007.30. PMC 2628762. PMID 18066065. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2628762.
- ^ Koscianska E. et al. (2007). "Prediction and preliminary validation of oncogene regulation by miRNAs". BMC Mol. Biol. 8: 79. doi:10.1186/1471-2199-8-79. PMC 2096627. PMID 17877811. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2096627.
- ^ Ioannidis P. et al. (2005). "CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells". J. Biol. Chem. 280 (20): 20086–20093. doi:10.1074/jbc.M410036200. PMID 15769738.
- ^ Johnson S.M. et al. (2005). "RAS is regulated by the let-7 microRNA family". Cell 120 (5): 635–647. doi:10.1016/j.cell.2005.01.014. PMID 15766527.
- ^ Mayr C. et al. (2007). "Disrupting the Pairing Between let-7 and Hmga2 Enhances Oncogenic Transformation". Science 315 (5818): 1576–1579. Bibcode 2007Sci...315.1576M. doi:10.1126/science.1137999. PMC 2556962. PMID 17322030. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2556962.
- ^ a b Johnson SM, Grosshans H, Shingara J et al. (2005). "Ras is regulated by the let-7 microrna family". Cell 120 (5): 635–47. doi:10.1016/j.cell.2005.01.014. PMID 15766527.
- ^ Johnson C.D. et al. (2007). "The let-7 microRNA represses cell proliferation pathways in human cells". Cancer Res. 67 (16): 7713–7722. doi:10.1158/0008-5472.CAN-07-1083. PMID 17699775.
- ^ He YJ, Guo L, D ZH. (2009) Let-7 and mir-24 in uvb-induced apoptosis [Chinese]. Zhonghua Fang She Yi Xue Yu Fang Hu Za Zhi. 29, 234–6.
- ^ Shell S, Park SM, Radjabi AR et al. (2007). "Let-7 expression defines two differentiation stages of cancer". Proc Natl Acad Sci U S A 104 (27): 11400–5. Bibcode 2007PNAS..10411400S. doi:10.1073/pnas.0704372104. PMC 2040910. PMID 17600087. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2040910.
- ^ a b Takamizawa J, Konishi H, Yanagisawa K et al. (2004). "Reduced expression of the let-7 micrornas in human lung cancers in association with shortened postoperative survival". Cancer Res. 64 (11): 3753–6. doi:10.1158/0008-5472.CAN-04-0637. PMID 15172979.
- ^ Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S (2007). "Role of Dicer and Drosha for endothelial microrna expression and angiogenesis". Circ Res 101 (1): 59–68. doi:10.1161/CIRCRESAHA.107.153916. PMID 17540974.
- ^ Esquela , Kerscher A, Trang P, Wiggins JF et al. (2008). "The let-7 microrna reduces tumor growth in mouse models of lung cancer". Cell Cycle 7 (6): 759–64. doi:10.4161/cc.7.6.5834. PMID 18344688.
- ^ Barh D., Malhotra R., Ravi B., and Sindhurani P. (2010). MicroRNA let-7: an emerging next-generation cancer therapeutic. CURRENT ONCOLOGY. 17, 70-80.
Further reading
- ^ Dangi-Garimella S, Strouch MJ, Grippo PJ, Bentrem DJ, Munshi HG (2010). "Collagen regulation of let-7 in pancreatic cancer involves TGF-β1-mediated membrane type 1-matrix metalloproteinase expression". Oncogene 30 (8): 1002–1008. doi:10.1038/onc.2010.485. PMC 3172057. PMID 21057545. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3172057.
- ^ Yang X, Lin X, Zhong X, Kaur S, Li N, Liang S, Lassus H, Wang L, Katsaros D, Montone K, Zhao X, Zhang Y, Bützow R, Coukos G, Zhang L (2010). "Double negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells". Cancer Res 70 (22): 9463–9472. doi:10.1158/0008-5472.CAN-10-2388. PMC 3057570. PMID 21045151. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3057570.
- ^ Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T (2010). Wölfl, Stefan. ed. "Let-7 MicroRNA Family Is Selectively Secreted into the Extracellular Environment via Exosomes in a Metastatic Gastric Cancer Cell Line". PLoS One 5 (10): e13247. doi:10.1371/journal.pone.0013247. PMC 2951912. PMID 20949044. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2951912.
- ^ Ramachandran R, Fausett BV, Goldman D (2010). "Ascl1a regulates Müller glia dedifferentiation and retina regeneration via a Lin-28-dependent, let-7 miRNA signaling pathway". Nat Cell Biol 12 (11): 1101–7. doi:10.1038/ncb2115. PMC 2972404. PMID 20935637. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2972404.
- ^ Ruzzo A, Canestrari E, Galluccio N, Santini D, Vincenzi B, Tonini G, Magnani M, Graziano F (2010). "Role of KRAS let-7 LCS6 SNP in metastatic colorectal cancer patients". Ann Oncol 22 (1): 234–5. doi:10.1093/annonc/mdq472. PMID 20926546.
- ^ Garbuzov A, Tatar M (2010). "Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity". Fly (Austin) 4 (4): 306–11. doi:10.4161/fly.4.4.13008. PMC 3174482. PMID 20798594. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3174482.
- ^ Ji J, Wang XW (2010). "A Yin-Yang Balancing Act of the Lin28/Let-7 Link in Tumorigenesis". J Hepatol 53 (5): 974–5. doi:10.1016/j.jhep.2010.07.001. PMC 2949515. PMID 20739081. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2949515.
- ^ Osada H, Takahashi T (2010). "Review Article: let-7 and miR-17-92: Small-sized major players in lung cancer development". Cancer Sci 102 (1): 9–17. doi:10.1111/j.1349-7006.2010.01707.x. PMID 20735434.
- ^ He Y, Yang C, Kirkmire CM, Wang ZJ (2010). "Regulation of opioid tolerance by let-7 family microRNA targeting the μ opioid receptor". J Neurosci 30 (30): 10251–8. doi:10.1523/JNEUROSCI.2419-10.2010. PMC 2943348. PMID 20668208. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2943348.
- ^ Cevec M, Thibaudeau C, Plavec J (2010). "NMR structure of the let-7 miRNA interacting with the site LCS1 of lin-41 mRNA from Caenorhabditis elegans". Nucleic Acids Res 38 (21): 7814–21. doi:10.1093/nar/gkq640. PMC 2995062. PMID 20660479. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2995062.
- ^ Nie K, Zhang T, Allawi H, Gomez M, Liu Y, Chadburn A, Wang YL, Knowles DM, Tam W (2010). "Epigenetic Down-Regulation of the Tumor Suppressor Gene PRDM1/Blimp-1 in Diffuse Large B Cell Lymphomas : A Potential Role of the MicroRNA Let-7". Am J Pathol 177 (3): 1470–9. doi:10.2353/ajpath.2010.091291. PMC 2928978. PMID 20651244. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2928978.
- ^ Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, Coarfa C, Zariff A, Milosavljevic A, Batts LM, Kheradmand F, Gunaratne PH, Corry DB (2010). "Proinflammatory Role for let-7 MicroRNAS in Experimental Asthma". J Biol Chem 285 (39): 30139–49. doi:10.1074/jbc.M110.145698. PMC 2943272. PMID 20630862. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2943272.
- ^ Newman MA, Hammond SM (2010). "Lin-28: an early embryonic sentinel that blocks Let-7 biogenesis". Int J Biochem Cell Biol 42 (8): 1330–3. doi:10.1016/j.biocel.2009.02.023. PMID 20619222.
- ^ Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim SK, Park CK, Jung KH, Lee SK, Kim M, Roh JK (2010). "Let-7 microRNA inhibits the proliferation of human glioblastoma cells". J Neurooncol 102 (1): 19–24. doi:10.1007/s11060-010-0286-6. PMID 20607356.
- ^ Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, Hu-Lieskovan S, Mauro DJ, Langer C, Rowinsky EK, Lenz HJ (2010). "A let-7 microRNA-binding site polymorphism in 3'-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy". Ann Oncol 22 (1): 104–9. doi:10.1093/annonc/mdq315. PMID 20603437.
- ^ Zhao Y, Deng C, Wang J, Xiao J, Gatalica Z, Recker RR, Xiao GG (2010). "Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer". Breast Cancer Res Treat 127 (1): 69–80. doi:10.1007/s10549-010-0972-2. PMID 20535543.
- ^ Hu G, Zhou R, Liu J, Gong AY, Chen XM (2010). "MicroRNA-98 and let-7 Regulate Expression of Suppressor of Cytokine Signaling-4 in Biliary Epithelial Cells in Response to Cryptosporidium parvum Infection". J Infect Dis 202 (1): 125–35. doi:10.1086/653212. PMC 2880649. PMID 20486857. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2880649.
- ^ Steinemann D, Tauscher M, Praulich I, Niemeyer CM, Flotho C, Schlegelberger B (2010). "Mutations in the let-7 binding site - a mechanism of RAS activation in juvenile myelomonocytic leukemia?". Haematologica 95 (9): 1616. doi:10.3324/haematol.2010.024984. PMC 2930968. PMID 20460640. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2930968.
- ^ Wong TS, Man OY, Tsang CM, Tsao SW, Tsang RK, Chan JY, Ho WK, Wei WI, To VS (2010). "MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression". J Cancer Res Clin Oncol 137 (3): 415–422. doi:10.1007/s00432-010-0898-4. PMC 3036828. PMID 20440510. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3036828.
- ^ Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H, Doki Y, Mori M, Hayashi N (2010). "The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma". J Hepatol 52 (5): 698–704. doi:10.1016/j.jhep.2009.12.024. PMID 20347499.
- ^ Jakymiw A, Patel RS, Deming N, Bhattacharyya I, Shah P, Lamont RJ, Stewart CM, Cohen DM, Chan EK (2010). "Overexpression of Dicer as a Result of Reduced let-7 microRNA Levels Contributes to Increased Cell Proliferation of Oral Cancer Cells". Genes Chromosomes Cancer 49 (6): 549–59. doi:10.1002/gcc.20765. PMC 2859695. PMID 20232482. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2859695.
- ^ Koh W, Sheng CT, Tan B, Lee QY, Kuznetsov V, Kiang LS, Tanavde V (2010). "Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of Hepatic Nuclear Factor 4 Alpha". BMC Genomics 11 Suppl 1: S6. doi:10.1186/1471-2164-11-S1-S6. PMC 2822534. PMID 20158877. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2822534.
- ^ Barh D, Malhotra R, Ravi B, Sindhurani P (2010). "Microrna let-7: an emerging next-generation cancer therapeutic". Curr Oncol 17 (1): 70–80. PMC 2826782. PMID 20179807. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2826782.
- ^ Balzer E, Heine C, Jiang Q, Lee VM, Moss EG (2010). "LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro". Development 137 (6): 891–900. doi:10.1242/dev.042895. PMID 20179095.
- ^ Graziano F, Canestrari E, Loupakis F, Ruzzo A, Galluccio N, Santini D, Rocchi M, Vincenzi B, Salvatore L, Cremolini C, Spoto C, Catalano V, D'Emidio S, Giordani P, Tonini G, Falcone A, Magnani M (2010). "Genetic modulation of the Let-7 microRNA binding to KRAS 3'-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan". Pharmacogenomics J 10 (5): 458–64. doi:10.1038/tpj.2010.9. PMID 20177422.
- ^ Klemke M, Meyer A, Hashemi Nezhad M, Belge G, Bartnitzke S, Bullerdiek J (2010). "Loss of let-7 binding sites resulting from truncations of the 3' untranslated region of HMGA2 mRNA in uterine leiomyomas". Cancer Genet Cytogenet 196 (2): 119–23. doi:10.1016/j.cancergencyto.2009.09.021. PMID 20082846.
- ^ Oh JS, Kim JJ, Byun JY, Kim IA (2010). "Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras". Int J Radiat Oncol Biol Phys 76 (1): 5–8. doi:10.1016/j.ijrobp.2009.08.028. PMID 20005451.
- ^ Mu G, Liu H, Zhou F, Xu X, Jiang H, Wang Y, Qu Y (2010). "Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 down-regulation in retinoblastomas". Hum Pathol 41 (4): 493–502. doi:10.1016/j.humpath.2009.08.022. PMID 20004941.
- ^ Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, Slack FJ (2010). "Regression of murine lung tumors by the let-7 microRNA". Oncogene 29 (11): 1580–7. doi:10.1038/onc.2009.445. PMC 2841713. PMID 19966857. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2841713.
- ^ Ricarte-Filho JC, Fuziwara CS, Yamashita AS, Rezende E, da-Silva MJ, Kimura ET (2009). "Effects of let-7 microRNA on Cell Growth and Differentiation of Papillary Thyroid Cancer". Transl Oncol 2 (4): 236–41. PMC 2781070. PMID 19956384. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2781070.
- ^ Noh SJ, Miller SH, Lee YT, Goh SH, Marincola FM, Stroncek DF, Reed C, Wang E, Miller JL (2009). "Let-7 microRNAs are developmentally regulated in circulating human erythroid cells". J Transl Med 7: 98. doi:10.1186/1479-5876-7-98. PMC 2792219. PMID 19939273. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2792219.
- ^ Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG (2009). "The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2". Nat Cell Biol 11 (12): 1411–20. doi:10.1038/ncb1987. PMID 19898466.
- ^ Iliopoulos D, Hirsch HA, Struhl K (2009). "An epigenetic switch involving NF-κB, Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation". Cell 139 (4): 693–706. doi:10.1016/j.cell.2009.10.014. PMC 2783826. PMID 19878981. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2783826.
- ^ Hammell CM, Karp X, Ambros V (2009). "A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans". Proc Natl Acad Sci U S A 106 (44): 18668–73. doi:10.1073/pnas.0908131106. PMC 2774035. PMID 19828440. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2774035.
- ^ Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME (2010). "The role of let-7 in cell differentiation and cancer". Endocr Relat Cancer 17 (1): F19–36. doi:10.1677/ERC-09-0184. PMID 19779035.
- ^ Hagan JP, Piskounova E, Gregory RI (2009). "Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in embryonic stem cells". Nat Struct Mol Biol 16 (10): 1021–5. doi:10.1038/nsmb.1676. PMC 2758923. PMID 19713958. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2758923.
- ^ Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA (2009). "LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans". Nat Struct Mol Biol 16 (10): 1016–20. doi:10.1038/nsmb.1675. PMC 2988485. PMID 19713957. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2988485.
- ^ Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH (2009). "Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells". Cancer Res 69 (16): 6704–12. doi:10.1158/0008-5472.CAN-09-1298. PMC 2727571. PMID 19654291. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2727571.
- ^ Roush SF, Slack FJ (2009). "Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1". Dev Biol 334 (2): 523–34. doi:10.1016/j.ydbio.2009.07.012. PMC 2753757. PMID 19627983. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2753757.
- ^ Chan SP, Slack FJ (2009). "Ribosomal protein RPS-14 modulates let-7 microRNA function in Caenorhabditis elegans". Dev Biol 334 (1): 152–60. doi:10.1016/j.ydbio.2009.07.011. PMC 2753218. PMID 19627982. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2753218.
- ^ Shi G, Perle MA, Mittal K, Chen H, Zou X, Narita M, Hernando E, Lee P, Wei JJ (2009). "Let-7 repression leads to HMGA2 overexpression in uterine leiomyosarcoma". J Cell Mol Med 13 (9B): 3898–905. doi:10.1111/j.1582-4934.2008.00541.x. PMID 19602040.
- ^ Hu G, Zhou R, Liu J, Gong AY, Eischeid AN, Dittman JW, Chen XM (2009). "MicroRNA-98 and let-7 Confer Cholangiocyte Expression of Cytokine-Inducible Src Homology 2-Containing Protein in Response to Microbial Challenge,". J Immunol 183 (3): 1617–24. doi:10.4049/jimmunol.0804362. PMC 2906382. PMID 19592657. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2906382.
- ^ Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009). "HuR recruits let-7/RISC to repress c-Myc expression". Genes Dev 23 (15): 1743–8. doi:10.1101/gad.1812509. PMC 2720259. PMID 19574298. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2720259.
- ^ Wang X, Hulshizer RL, Erickson-Johnson MR, Flynn HC, Jenkins RB, Lloyd RV, Oliveira AM (2009). "Identification of novel HMGA2 fusion sequences in lipoma: evidence that deletion of let-7 miRNA consensus binding site 1 in the HMGA2 3' UTR is not critical for HMGA2 transcriptional upregulation". Genes Chromosomes Cancer 48 (8): 673–8. doi:10.1002/gcc.20674. PMID 19431195.
- ^ Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, Marsit CJ, Kelsey KT (2009). "A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers". Carcinogenesis 30 (6): 1003–7. doi:10.1093/carcin/bgp099. PMC 2691138. PMID 19380522. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2691138.
- ^ Slack F (2009). "let-7 microRNA reduces tumor growth". Cell Cycle 8 (12): 1823. PMID 19377282.
- ^ Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ (2009). "MicroRNA let-7 Regulates 3T3-L1 Adipogenesis". Mol Endocrinol 23 (6): 925–31. doi:10.1210/me.2008-0298. PMC 2691679. PMID 19324969. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2691679.
- ^ Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, Buscail L, Cordelier P (2009). "let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression". Hum Gene Ther 20 (8): 831–44. doi:10.1089/hum.2008.134. PMID 19323605.
- ^ Peter ME (2009). "Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression". Cell Cycle 8 (6): 843–52. doi:10.4161/cc.8.6.7907. PMC 2688687. PMID 19221491. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2688687.
- ^ Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT (2009). "Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation". Proc Natl Acad Sci U S A 106 (9): 3384–9. Bibcode 2009PNAS..106.3384C. doi:10.1073/pnas.0808300106. PMC 2651245. PMID 19211792. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2651245.
- ^ Rahman MM, Qian ZR, Wang EL, Sultana R, Kudo E, Nakasono M, Hayashi T, Kakiuchi S, Sano T (2009). "Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation". Br J Cancer 100 (3): 501–10. doi:10.1038/sj.bjc.6604883. PMC 2658538. PMID 19156147. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2658538.
- ^ Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR (2009). "Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7". EMBO J 28 (4): 347–58. doi:10.1038/emboj.2008.294. PMC 2646152. PMID 19153603. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2646152.
- ^ Qian ZR, Asa SL, Siomi H, Siomi MC, Yoshimoto K, Yamada S, Wang EL, Rahman MM, Inoue H, Itakura M, Kudo E, Sano T (2009). "Overexpression of HMGA2 relates to reduction of the let-7 and its relationship to clinicopathological features in pituitary adenomas". Mod Pathol 22 (3): 431–41. doi:10.1038/modpathol.2008.202. PMID 19136928.
- ^ Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA (2009). "let-7 Overexpression Leads to an Increased Fraction of Cells in G2/M, Direct Down-regulation of Cdc34, and Stabilization of Wee1 Kinase in Primary Fibroblasts". J Biol Chem 284 (11): 6605–9. doi:10.1074/jbc.C900002200. PMC 2652271. PMID 19126550. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2652271.
- ^ Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ (2008). "The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure". Cell Cycle 7 (24): 3935–42. doi:10.4161/cc.7.24.7397. PMC 2895810. PMID 19098426. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2895810.
- ^ Heo I, Joo C, Cho J, Ha M, Han J, Kim VN (2008). "Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA". Mol Cell 32 (2): 276–84. doi:10.1016/j.molcel.2008.09.014. PMID 18951094.
- ^ Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB (2008). "A SNP in a let-7 microRNA complementary site in the KRAS 3′UTR Increases Non-Small Cell Lung Cancer Risk". Cancer Res 68 (20): 8535–40. doi:10.1158/0008-5472.CAN-08-2129. PMC 2672193. PMID 18922928. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2672193.
- ^ Andachi Y (2008). "A novel biochemical method to identify target genes of individual microRNAs: Identification of a new Caenorhabditis elegans let-7 target". RNA 14 (11): 2440–51. doi:10.1261/rna.1139508. PMC 2578851. PMID 18824511. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2578851.
- ^ Ding XC, Slack FJ, Grosshans H (2008). "The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation". Cell Cycle 7 (19): 3083–90. doi:10.4161/cc.7.19.6778. PMC 2887667. PMID 18818519. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2887667.
- ^ Forman JJ, Legesse-Miller A, Coller HA (2008). "A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence". Proc Natl Acad Sci U S A 105 (39): 14879–84. Bibcode 2008PNAS..10514879F. doi:10.1073/pnas.0803230105. PMC 2567461. PMID 18812516. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2567461.
- ^ Roush S, Slack FJ (2008). "The let-7 family of microRNAs". Trends Cell Biol 18 (10): 505–16. doi:10.1016/j.tcb.2008.07.007. PMID 18774294.
- ^ Tennessen JM, Thummel CS (2008). "let-7: Developmental timing conserved through evolution". Curr Biol 18 (16): R707–8. doi:10.1016/j.cub.2008.07.013. PMC 2583239. PMID 18727906. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2583239.
- ^ Chan SP, Ramaswamy G, Choi EY, Slack FJ (2008). "Identification of specific let-7 microRNA binding complexes in Caenorhabditis elegans". RNA 14 (10): 2104–14. doi:10.1261/rna.551208. PMC 2553747. PMID 18719242. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2553747.
- ^ Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T (2008). "let-7 regulates Dicer expression and constitutes a negative feedback loop". Carcinogenesis 29 (11): 2073–7. doi:10.1093/carcin/bgn187. PMID 18700235.
- ^ Büssing I, Slack FJ, Grosshans H (2008). "let-7 microRNAs in development, stem cells and cancer". Trends Mol Med 14 (9): 400–9. doi:10.1016/j.molmed.2008.07.001. PMID 18674967.
- ^ Jérôme T, Laurie P, Louis B, Pierre C (2007). "Enjoy the Silence: The Story of let-7 MicroRNA and Cancer". Curr Genomics 8 (4): 229–33. doi:10.2174/138920207781386933. PMC 2430685. PMID 18645597. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2430685.
- ^ Reid JG, Nagaraja AK, Lynn FC, Drabek RB, Muzny DM, Shaw CA, Weiss MK, Naghavi AO, Khan M, Zhu H, Tennakoon J, Gunaratne GH, Corry DB, Miller J, McManus MT, German MS, Gibbs RA, Matzuk MM, Gunaratne PH (2008). "Mouse let-7 miRNA populations exhibit RNA editing that is constrained in the 5′-seed/ cleavage/anchor regions and stabilize predicted mmu-let-7a:mRNA duplexes". Genome Res 18 (10): 1571–81. doi:10.1101/gr.078246.108. PMC 2556275. PMID 18614752. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2556275.
- ^ Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG (2008). "A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment". Nat Cell Biol 10 (8): 987–93. doi:10.1038/ncb1759. PMID 18604195.
- ^ Caygill EE, Johnston LA (2008). "Temporal Regulation of Metamorphic Processes in Drosophila by the let-7 and miR-125 Heterochronic MicroRNAs". Curr Biol 18 (13): 943–50. doi:10.1016/j.cub.2008.06.020. PMC 2736146. PMID 18571409. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2736146.
- ^ Newman MA, Thomson JM, Hammond SM (2008). "Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing". RNA 14 (8): 1539–49. doi:10.1261/rna.1155108. PMC 2491462. PMID 18566191. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2491462.
- ^ Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H, Bell JC (2008). "A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication". Mol Ther 16 (8): 1437–43. doi:10.1038/mt.2008.130. PMID 18560417.
- ^ Sokol NS, Xu P, Jan YN, Ambros V (2008). "Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis". Genes Dev 22 (12): 1591–6. doi:10.1101/gad.1671708. PMC 2428057. PMID 18559475. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2428057.
- ^ Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M (2008). "Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family". Clin Cancer Res 14 (8): 2334–40. doi:10.1158/1078-0432.CCR-07-4667. PMID 18413822.
- ^ Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME (2008). "Identification of let-7-regulated oncofetal genes". Cancer Res 68 (8): 2587–91. doi:10.1158/0008-5472.CAN-08-0264. PMID 18413726.
- ^ Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, Wei JJ (2008). "Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma". Mol Cancer Res 6 (4): 663–73. doi:10.1158/1541-7786.MCR-07-0370. PMID 18403645.
- ^ Garfield D (2008). "let-7 microRNA expression and the distinction between nonmucinous and mucinous bronchioloalveolar carcinomas". Lung Cancer 60 (2): 307. doi:10.1016/j.lungcan.2008.02.010. PMID 18395292.
- ^ Dröge P, Davey CA (2008). "Do cells let-7 determine stemness?". Cell Stem Cell 2 (1): 8–9. doi:10.1016/j.stem.2007.12.003. PMID 18371414.
- ^ Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ (2008). "The let-7 microRNA reduces tumor growth in mouse models of lung cancer". Cell Cycle 7 (6): 759–64. doi:10.4161/cc.7.6.5834. PMID 18344688.
- ^ Solomon A, Mian Y, Ortega-Cava C, Liu VW, Gurumurthy CB, Naramura M, Band V, Band H (2008). "Upregulation of the let-7 microRNA with precocious development in lin-12/Notch hypermorphic C. elegans mutants". Dev Biol 316 (2): 191–9. doi:10.1016/j.ydbio.2007.12.046. PMC 2390880. PMID 18334253. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2390880.
- ^ Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T (2008). "Suppression of non-small cell lung tumor development by the let-7 microRNA family". Proc Natl Acad Sci U S A 105 (10): 3903–8. Bibcode 2008PNAS..105.3903K. doi:10.1073/pnas.0712321105. PMC 2268826. PMID 18308936. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2268826.
- ^ Cevec M, Thibaudeau C, Plavec J (2008). "Solution structure of a let-7 miRNA:lin-41 mRNA complex from C. elegans". Nucleic Acids Res 36 (7): 2330–7. doi:10.1093/nar/gkn088. PMC 2367737. PMID 18296482. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2367737.
- ^ Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E (2007). "let-7 regulates self renewal and tumorigenicity of breast cancer cells". Cell 131 (6): 1109–23. doi:10.1016/j.cell.2007.10.054. PMID 18083101.
- ^ O'Farrell F, Esfahani SS, Engström Y, Kylsten P (2008). "Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade". Dev Dyn 237 (1): 196–208. doi:10.1002/dvdy.21396. PMID 18069688.
- ^ Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME (2007). "Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2". Cell Cycle 6 (21): 2585–90. doi:10.4161/cc.6.21.4845. PMID 17957144.
- ^ Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH (2007). "Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development". Mol Biol Evol 24 (11): 2525–34. doi:10.1093/molbev/msm195. PMID 17890240.
- ^ Tsonis PA, Call MK, Grogg MW, Sartor MA, Taylor RR, Forge A, Fyffe R, Goldenberg R, Cowper-Sal-lari R, Tomlinson CR (2007). "microRNAs and regeneration: let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt". Biochem Biophys Res Commun 362 (4): 940–5. doi:10.1016/j.bbrc.2007.08.077. PMC 2683343. PMID 17765873. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2683343.
- ^ Inamura K, Togashi Y, Nomura K, Ninomiya H, Hiramatsu M, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y (2007). "let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis". Lung Cancer 58 (3): 392–6. doi:10.1016/j.lungcan.2007.07.013. PMID 17728006.
- ^ Salzman DW, Shubert-Coleman J, Furneaux H (2007). "P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression". J Biol Chem 282 (45): 32773–9. doi:10.1074/jbc.M705054200. PMID 17724023.
- ^ Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ (2007). "The let-7 microRNA represses cell proliferation pathways in human cells". Cancer Res 67 (16): 7713–22. doi:10.1158/0008-5472.CAN-07-1083. PMID 17699775.
- ^ Wakiyama M, Takimoto K, Ohara O, Yokoyama S (2007). "Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system". Genes Dev 21 (15): 1857–62. doi:10.1101/gad.1566707. PMC 1935024. PMID 17671087. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1935024.
- ^ Liu S, Xia Q, Zhao P, Cheng T, Hong K, Xiang Z (2007). "Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori)". BMC Dev Biol 7: 88. doi:10.1186/1471-213X-7-88. PMC 1976426. PMID 17651473. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1976426.
- ^ Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME (2007). "Let-7 expression defines two differentiation stages of cancer". Proc Natl Acad Sci U S A 104 (27): 11400–5. Bibcode 2007PNAS..10411400S. doi:10.1073/pnas.0704372104. PMC 2040910. PMID 17600087. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2040910.
- ^ Lee YS, Dutta A (2007). "The tumor suppressor microRNA let-7 represses the HMGA2 oncogene". Genes Dev 21 (9): 1025–30. doi:10.1101/gad.1540407. PMC 1855228. PMID 17437991. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1855228.
- ^ Nolde MJ, Saka N, Reinert KL, Slack FJ (2007). "The C. elegans pumilio homolog, puf-9, is required for the 3'UTR mediated repression of the let-7 microRNA target gene, hbl-1". Dev Biol 305 (2): 551–63. doi:10.1016/j.ydbio.2007.02.040. PMC 2096746. PMID 17412319. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2096746.
- ^ Hayes GD, Ruvkun G (2006). "Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs". Cold Spring Harb Symp Quant Biol 71: 21–7. doi:10.1101/sqb.2006.71.018. PMID 17381276.
- ^ Mayr C, Hemann MT, Bartel DP (2007). "Disrupting the Pairing Between let-7 and Hmga2 Enhances Oncogenic Transformation". Science 315 (5818): 1576–9. Bibcode 2007Sci...315.1576M. doi:10.1126/science.1137999. PMC 2556962. PMID 17322030. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2556962.
- ^ Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R (2007). "Post-transcriptional regulation of the let-7 microRNA during neural cell specification". FASEB J 21 (2): 415–26. doi:10.1096/fj.06-6130com. PMID 17167072.
- ^ Hayes GD, Frand AR, Ruvkun G (2006). "The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25". Development 133 (23): 4631–41. doi:10.1242/dev.02655. PMID 17065234.
- ^ Akao Y, Nakagawa Y, Naoe T (2006). "let-7 microRNA functions as a potential growth suppressor in human colon cancer cells". Biol Pharm Bull 29 (5): 903–6. doi:10.1248/bpb.29.903. PMID 16651716.
- ^ Schulman BR, Esquela-Kerscher A, Slack FJ (2005). "Reciprocal Expression of lin-41 and the microRNAs let-7 and mir-125 During Mouse Embryogenesis". Dev Dyn 234 (4): 1046–54. doi:10.1002/dvdy.20599. PMC 2596717. PMID 16247770. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2596717.
- ^ Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ (2005). "Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system". Dev Dyn 234 (4): 868–77. doi:10.1002/dvdy.20572. PMC 2572564. PMID 16217741. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2572564.
- ^ Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE (2005). "Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects". Dev Cell 9 (3): 415–22. doi:10.1016/j.devcel.2005.08.002. PMID 16139229.
- ^ Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V (2005). "The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans". Dev Cell 9 (3): 403–14. doi:10.1016/j.devcel.2005.07.009. PMID 16139228.
- ^ Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE (2005). "Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation". Cell 122 (4): 553–63. doi:10.1016/j.cell.2005.07.031. PMID 16122423.
- ^ Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W (2005). "Inhibition of translational initiation by Let-7 MicroRNA in human cells". Science 309 (5740): 1573–6. Bibcode 2005Sci...309.1573P. doi:10.1126/science.1115079. PMID 16081698.
- ^ Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005). "RAS is regulated by the let-7 microRNA family". Cell 120 (5): 635–47. doi:10.1016/j.cell.2005.01.014. PMID 15766527.
- ^ Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ (2005). "The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans". Dev Cell 8 (3): 321–30. doi:10.1016/j.devcel.2004.12.019. PMID 15737928.
- ^ Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH (2004). "Substrate requirements for let-7 function in the developing zebrafish embryo". Nucleic Acids Res 32 (21): 6284–91. doi:10.1093/nar/gkh968. PMC 535676. PMID 15585662. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=535676.
- ^ Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE (2004). "Trans-splicing and polyadenylation of let-7 microRNA primary transcripts". RNA 10 (10): 1586–94. doi:10.1261/rna.7122604. PMC 1370645. PMID 15337850. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1370645.
- ^ Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T (2004). "Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival". Cancer Res 64 (11): 3753–6. doi:10.1158/0008-5472.CAN-04-0637. PMID 15172979.
- ^ Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004). "The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR". Genes Dev 18 (2): 132–7. doi:10.1101/gad.1165404. PMC 324419. PMID 14729570. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=324419.
- ^ Basyuk E, Suavet F, Doglio A, Bordonné R, Bertrand E (2003). "Human let-7 stem–loop precursors harbor features of RNase III cleavage products". Nucleic Acids Res 31 (22): 6593–7. doi:10.1093/nar/gkg855. PMC 275551. PMID 14602919. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=275551.
- ^ Johnson SM, Lin SY, Slack FJ (2003). "The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter". Dev Biol 259 (2): 364–79. doi:10.1016/S0012-1606(03)00202-1. PMID 12871707.
- ^ Pasquinelli AE, McCoy A, Jiménez E, Saló E, Ruvkun G, Martindale MQ, Baguñà J (2003). "Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution?". Evol Dev 5 (4): 372–8. doi:10.1046/j.1525-142X.2003.03044.x. PMID 12823453.
- ^ Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V (2002). "The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster". Dev Biol 244 (1): 170–9. doi:10.1006/dbio.2002.0594. PMID 11900466.
- ^ Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD (2001). "A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA". Science 293 (5531): 834–8. doi:10.1126/science.1062961. PMID 11452083.
- ^ Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000). "Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA". Nature 408 (6808): 86–9. doi:10.1038/35040556. PMID 11081512.
- ^ Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G (2000). "The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor". Mol Cell 5 (4): 659–69. doi:10.1016/S1097-2765(00)80245-2. PMID 10882102.
- ^ Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000). "The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans". Nature 403 (6772): 901–6. Bibcode 2000Natur.403..901R. doi:10.1038/35002607. PMID 10706289.
External links
miRNA precursor families 1-100 101-200 201+ Other Categories:
Wikimedia Foundation. 2010.
