- Protein moonlighting
-
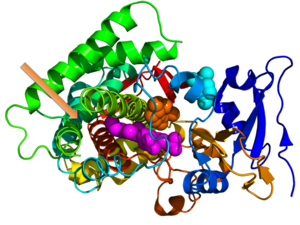 Crystallographic structure of cytochrome P450 from the bacteria S. coelicolor (rainbow colored cartoon, N-terminus = blue, C-terminus = red) complexed with heme cofactor (magenta spheres) and two molecules of its endogenous substrate epi-isozizaene as orange and cyan spheres respectively. The orange-colored substrate resides in the monooxygenase site while the cyan-colored substrate occupies the substrate entrance site. An unoccupied moonlighting terpene synthase site is designated by the orange arrow.[1]
Crystallographic structure of cytochrome P450 from the bacteria S. coelicolor (rainbow colored cartoon, N-terminus = blue, C-terminus = red) complexed with heme cofactor (magenta spheres) and two molecules of its endogenous substrate epi-isozizaene as orange and cyan spheres respectively. The orange-colored substrate resides in the monooxygenase site while the cyan-colored substrate occupies the substrate entrance site. An unoccupied moonlighting terpene synthase site is designated by the orange arrow.[1] This article is about genes sharing multiple functions in an organism. For "gene sharing" between organisms, see horizontal gene transfer.
This article is about genes sharing multiple functions in an organism. For "gene sharing" between organisms, see horizontal gene transfer.Protein moonlighting (or gene sharing) is a phenomenon by which a protein can perform more than one function.[2] Ancestral moonlighting proteins originally possessed a single function but through evolution, acquired additional functions. Many proteins that moonlight are enzymes; others are receptors, ion channels or chaperones. The most common primary function of moonlighting proteins is enzymatic catalysis, but these enzymes have acquired secondary non-enyzmatic roles. Some examples of functions of moonlighting proteins secondary to catalysis include signal transduction, transcriptional regulation, apoptosis, motility, and structural.[3]
Protein moonlighting may occur widely in nature. Protein moonlighting through gene sharing differs from the use of a single gene to generate different protein by alternative RNA splicing, DNA rearrangement, or post-translational processing. It is also different from multifunctionality of the protein, in which the protein has multiple domains, each serving a different function. Protein moonlighting by gene sharing means that a gene may acquire and maintain a second function without gene duplication and without loss of the primary function. Such genes are under two or more entirely different selective constraints.[4]
Various techniques have been used to reveal moonlighting functions in proteins. The detection of a protein in unexpected locations within cells, cell types, or tissues may suggest that a protein has a moonlighting function. Furthermore, sequence or structure homology of a protein may be used to infer both primary function as well as secondary moonlighting functions of a protein.
The most well-studied examples of gene sharing are crystallins. These proteins, when expressed at low levels in many tissues function as enzymes, but when expressed at high levels in eye tissue, become densely packed and thus form lenses. While the recognition of gene sharing is relatively recent—the term was coined in 1988, after crystallins in chickens and ducks were found to be identical to separately identified enzymes—recent studies have found many examples throughout the living world. Joram Piatigorsky has suggested that many or all proteins exhibit gene sharing to some extent, and that gene sharing is a key aspect of molecular evolution.[5]:1–7 The genes encoding crystallins must maintain sequences for catalytic function and transparency maintenance function.[4]
Inappropriate moonlighting is a contributing factor in some genetic diseases, and moonlighting provides a possible mechanism by which bacteria may become resistant to antibiotics.[6]
Contents
Discovery
The first observation of a moonlighting protein was made in the late 1980s by Joram Piatigorsky and Graeme Wistow during their research of crystallin enzymes. Piatigorsk determined that lens crystallins' conservation and variance is due to other moonlighting functions outside of the lens.[7] Originally Piatigorsky called these proteins "gene sharing" proteins, but the colloquial description moonlighting was subsequently applied to proteins by Constance Jeffery in 1999[8] to draw a similarity between multitasking proteins and people who work two jobs.[9] The phrase "gene sharing" is ambiguous since it is also used to describe horizontal gene transfer, hence the phrase "protein moonlighting" has become the preferred description for proteins with more than one function.[9]
Evolution
It is believed that moonlighting proteins came about by means of evolution through which uni-functional proteins gained the ability to perform multiple functions. With alterations, much of the protein's unused space can provide new functions.[6] Many moonlighting proteins are the result of gene fusion of two single function genes.[10] Alternatively a single gene can acquire a second function since the active site of the encoded protein typically is small compared to the overall size of the protein leaving considerable room to accommodate a second functional site. In yet a third alternative, the same active site can acquire a second function through mutations of the active site.
The development of moonlighting proteins may be evolutionary favorable to the organism since a single protein can do the job of multiple proteins conserving amino acids and energy required to synthesize these proteins.[8] However there is no universally agreed upon theory that explains why proteins with multiple roles evolved.[9][8] While using one protein to perform multiple roles seems advantageous because it keeps the genome small, we can conclude that this is probably not the reason for moonlighting because of the large of amount of noncoding DNA.[9]
Functions
Many proteins catalyze a chemical reaction. Other proteins fulfill structural, transport, or signaling roles. Furthermore, numerous proteins have the ability to aggregate into supramolecular assemblies. For example, a ribosome is made up of 90 proteins and RNA.
A number of the currently known moonlighting proteins are evolutionarily derived from highly conserved enzymes, also called ancient enzymes. These enzymes are frequently speculated to have evolved moonlighting functions. Since highly conserved proteins are present in many different organisms, this increases the chance that they would develop secondary moonlighting functions.[9] A high fraction of enzymes involved in glycolysis, an ancient universal metabolic pathway, exhibit moonlighting behavior. Furthermore it has been suggested that as many as 7 out of 10 proteins in glycolysis and 7 out of 8 enzymes of the tricarboxylic acid cycle exhibit moonlighting behavior.[3]
An example of a moonlighting enzyme is pyruvate carboxylase. This enzyme catalyzes the carboxylation of pyruvate into oxaloacetate, thereby replenishing the tricarboxylic acid cycle. Surprisingly, in yeast species such as H. polymorpha and P. pastoris, pyruvate carboylase is also essential for proper targeting and assembly of the peroxisomal protein alcohol oxidase (AO). AO, the first enzyme of methanol metabolism, is a homo-octameric flavoenzyme. In wild type cells, this enzyme is present as enzymatically active AO octamers in the peroxisomal matrix. However, in cells lacking pruvate carboxylase, AO monomers accumulate in the cytosol, indicating that pyruvate carboxylase has a second fully unrelated function in assembly and import. The function in AO import/assembly is fully independent of the enzyme activity of pyruvate carboxylase, because amino acid substitutions can be introduced that fully inactive the enzyme activity of pryuvate carboxylase, without affecting its function in AO assembly and import. Conversely, mutations are known that block the function of this enzyme in import and assembly of AO, but have no effect on the enzymatic activity of the protein.[9]
The E. coli anti-oxidant thioredoxin protein is another example of a moonlighting protein. Upon infection with the bacteriophage T7, E. coli thioredoxin forms a complex with T7 DNA polymerase, which results in enhanced T7 DNA replication, a crucial step for successful T7 infection. Thioredoxin binds to a loop in T7 DNA polymerase to bind more strongly to the DNA. The anti-oxidant function of thioredoxin is fully autonomous and fully independent of T7 DNA replication, in which the protein most likely fulfills the functional role.[9]
Examples of moonlighting proteins[9] Kingdom Protein Organism Function primary moonlighting Animal Aconitase H. sapiens TCA cycle enzyme Iron homeostasis ATF2 H. sapiens Transcription factor DNA damage respons Crystallins Various Lens structural protein Various enzyme Cytochrome c Various Energy metabolism Apoptosis DLD H. sapiens Energy metabolism Protease ERK2 H. sapiens MAP kinase Transcriptional repressor ESCRT-II complex D. melanogaster Endosomal protein sorting Biocoid mRNA localization STAT3 M. musculus Transcription factor Electron transport chain Plant Hexokinase A. thaliana Glucose metabolism Glucose signaling Presenilin P. patens γ-secretase Cystoskeletal function Fungus Aconitase S. cerevisiae TCA cycle enzyme mtDNA stability Aldolase S. cerevisiae Glycolytic enzyme V-ATPase assembly Arg5,6 S. cerevisiae Arginine biosynthesis Transcriptional control Enolase S. cerevisiae Glycolytic enzyme - Homotypic vacuole fusion
- Mitochondrial tRNA import
Galactokinase K. lactis Galactose catabolism enyzme Induction galactose genes Hal3 S. cerevisiae Halotolerance determinant Coenzyme A biosynthesis HSP60 S. cerevisiae Mitochondrial chaperone Stabilization active DNA ori's Phosphofructokinase P. pastoris Glycolytic enzyme Autophagy peroxisomes Pyruvate carboxylase H. polymorpha Anaplerotic enzyme Assembly of alcohol oxidase Vhs3 S. cerevisiae Halotolerance determinant Coenzyme A biosynthesis Prokaryotes Aconitase M. tuberculosis TCA cycle enzyme Iron-responsive protein CYP170A1 S. coelicolor Albaflavenone synthase Terpene synthase Enolase S. pneumoniae Glycolytic enzyme Plasminogen binding GroEL E. aerogenes Chaperone Insect toxin Glutamate racemase (MurI) E. coli cell wall biosynthesis gyrase inhibition Thioredoxin E. coli Anti-oxidant T7 DNA polymerase subunit Protist Aldolase P. vivax Glycolytic enzyme Host-cell invasion Mechanisms
In many cases, the functionality of a protein not only depends on its structure, but also its location. For example, a single protein may have one function when found in the cytoplasm of a cell, a different function when interacting with a membrane, and yet a third function if excreted from the cell. This property of moonlighting proteins is known as "differential localization".[12] For example, in higher temperatures DegP (HtrA) will function as a protease by the directed degradation of proteins and in lower temperatures as a chaperone by assisting the non-covalent folding or unfolding and the assembly or disassembly of other macromolecular structures.[6] Furthermore, moonlighting proteins may exhibit different behaviors not only as a result of its location within a cell, but also the type of cell that the protein is expressed in.[12]
Other methods through which proteins may moonlight are by changing their oligomeric state, altering concentrations of the protein's ligand or substrate, use of alternative binding sites, or finally through phosphorylation. An example of a protein that displays different function in different oligomeric states is pyruvate kinase which exhibits metabolic activity as a tetramer and thyroid hormone–binding activity as a monomer. Changes in the concentrations of ligands or substrates may cause a switch in protein a protein's function. For example, in the presence of low iron concentrations, aconitase functions as an enzyme while at high iron concentration, aconitase functions as an iron-responsive element-binding protein (IREBP). Proteins may also perform separate functions through the use of alternative binding sites that perform different tasks. An example of this is ceruloplasmin, a protein that functions as an oxidase in copper metabolism and moonlights as a copper-independent glutathione peroxidase. Lastly, phosphorylation may sometimes cause a switch in the function of a moonlighting protein. For example, phosphorylation of phosphoglucose isomerase (PGI) at Ser-185 by protein kinase CK2 causes it to stop functioning as an enzyme, while retaining its function as an autocrine motility factor.[3] Hence when a mutation takes place that inactivates a function of a moonlighting proteins, the other function(s) are not necessarily affected.[9]
The crystal structures of several moonlighting proteins, such as I-AniI homing endonuclease / maturase[13] and the PutA proline dehydrogenase / transcription factor,[14] have been determined.[15] An analysis of these crystal structures has demonstrated that moonlighting proteins can either perform both functions at the same time, or through conformational changes, alternate between two states, each of which is able to perform a separate function. For example, the protein DegP plays a role in proteolysis with higher temperatures and is involved in refolding functions at lower temperatures.[15] Lastly, these crystal structures have shown that the second function may negatively affect the first function in some moonlighting proteins. As seen in ƞ-crystallin, the second function of a protein can alter the structure, decreasing the flexibility, which in turn can impair enzymatic activity somewhat.[15]
Identification methods
Moonlighting proteins have usually been identified by chance because there is no clear procedure to identify secondary moonlighting functions. Despite such difficulties, the number of moonlighting proteins that have been discovered is rapidly increasing. Furthermore, moonlighting proteins appear to be abundant in all kingdoms of life.[9]
Various methods have been employed to determine a protein's function including secondary moonlighting functions. For example, the tissue, cellular, or subcellular distribution of a protein may provide hints as to the function. Real-time PCR is used to quantify mRNA and hence infer the presence or absence of a particular protein which is encoded by the mRNA within different cell types. Alternatively immunohistochemistry or mass spectrometry can be used to directly detect the presence of proteins and determine in which subcellular locations, cell types, and tissues a particular protein is expressed.
Mass spectrometry may be used to detect proteins based on their mass-to-charge ratio. Because of alternative splicing and posttranslational modification, identification of proteins based on the mass of the parent ion alone is very difficult. However tandem mass spectrometry in which each of the parent peaks is in turn fragmented can be used to unambiguously identify proteins. Hence tandem mass spectrometry is one of the tools used in proteomics to identify the presence of proteins in different cell types or subcellular locations. While the presence of a moonlighting protein in an unexpected location may complicate routine analyses, at the same time, the detection of a protein in unexpected multiprotein complexes or locations suggests that protein may have a moonlighting function.[12] Furthermore, mass spectrometry may be used to determine if a protein has high expression levels that do not correlate to the enzyme's measured metabolic activity. These expression levels may signify that the protein is performing a different function than previously known.[3]
The structure of a protein can also help determine its functions. Protein structure in turn may be elucidated with various techniques including X-ray crystallography or NMR. Dual polarization interferometry may be used to measure changes in protein structure which may also give hints to the protein's function. Finally, application of systems biology approaches[16] such as interactomics give clues to a proteins function based on what it interacts with.
Crystallins
 A crystallin from ducks that exhibits argininosuccinate lyase activity and is a key structural component in eye lenses, an example of gene sharing
A crystallin from ducks that exhibits argininosuccinate lyase activity and is a key structural component in eye lenses, an example of gene sharing
In the case of crystallins, the genes must maintain sequences for catalytic function and transparency maintenance function.[4] The abundant lens crystallins have been generally viewed as static proteins serving a strictly structural role in transparency and cataract.[17] However, recent studies have shown that the lens crystallins are much more diverse than previously recognized and that many are related or identical to metabolic enzymes and stress proteins found in numerous tissues.[18] Unlike other proteins performing highly specialized tasks, such as globin or rhodopsin, the crystallins are very diverse and show numerous species differences. Essentially all vertebrate lenses contain representatives of the α and β/γ crystallins, the "Ubiquitous crystallins", which are themselves heterogeneous, and only few species or selected taxonomic groups use entirely different proteins as lens crystallins.This paradox of crystallins being highly conserved in sequence while extremely diverse in number and distribution shows that many crystallins have vital functions outside the lens and cornea, and this multi-functionality of the crystallins is achieved by gene sharing.[19]
Gene regulation
Crystallin recruitment may occur by changes in gene regulation that leads to high lens expression. One such example is gluthathione S-transferase/S11-crystallin that was specialized for lens expression by change in gene regulation and gene duplication. The fact that similar transcriptional factors such as Pax-6, and retinoic acid receptors, regulate different crystalline genes, suggests that lens-specific expression have played a crucial role for recruiting multifunctional protein as crystallins. Crystallin recruitment has occurred both with and without gene duplication, and tandem gene duplication has taken place among some of the crystallins with one of the duplicates specializing for lens expression. Ubiquitous α –crystallins and bird δ –crystallins are two examples.[20]
Alpha crystallins
The α-crystallins, which contributed to the discovery of crystallins as borrowed proteins,[21] have continually supported the theory of gene sharing, and helped delineating the mechanisms used for gene sharing as well. There are two α-crystallin genes (αA and αB), which are about 55% identical in amino acid sequence.[18] Expression studies in non-lens cells showed that the αB-crystallin, other than being a functional lens protein, is a functional small heat shock protein.[22] αB-crystallin is induced by heat and other physiological stresses, and it can protect the cells from elevated temperatures[23] and hypertonic stress.[24] αB-crystallin is also overexpressed in many pathologies, including neurodegenerative diseases, fibroblasts of patients with Werner's disease showing premature senescence, and growth abnormalities. In addition to being overexpressed under abnormal conditions, αB-crystallin is constitutively expressed in heart, skeletal muscle, kidney, lung and many other tissues.[25] In contrast to αB-crystallin, except for low-level expression in the thymus, spleen and retina,[26] αA-crystallin is highly specialized for expression in the lens[27] and is not stress-inducible. However, like αB-crystallin, it can also function as molecular chaperone and protect against thermal stress.
Beta/gamma-crystallins
β/γ-crystallins are different from α-crystallins in that they are a large multigene family. Other proteins like bacterial spore coat, a slime mold cyst protein, and epidermis differentiation-specific protein, contain the same Greek key motifs and are placed under β/γ crystallin superfamily. This relationship supports the idea that β/γ- crystallins have been recruited by a gene-sharing mechanism. However, except for few reports, non-refractive function of the β/γ-crystallin is yet to be found.[19]
Corneal crystallins
Similar to lens, cornea is a transparent, avascular tissue derived from the ectoderm that is responsible for focusing light onto the retina. However, unlike lens, cornea depends on the air-cell interface and its curvature for refraction. Early immunology studies have shown that BCP 54 comprises 20–40% of the total soluble protein in bovine cornea.[28] Subsequent studies have indicated that BCP 54 is ALDH3, a tumor and xenobiotic-inducible cytosolic enzyme, found in human, rat, and other mammals.[29]
Non refractive roles of crystallins in lens and cornea
While it is evident that gene sharing resulted in many of lens crystallins being multifunctional proteins, it is still uncertain to what extent the crystallins use their non-refractive properties in the lens, or on what basis they were selected. The α-crystallins provide a convincing case for a lens crystallin using its non-refractive ability within the lens to prevent protein aggregation under a variety of environmental stresses[30] and to protect against enzyme inactivation by post-translational modifications such as glycation.[31] The α-crystallins may also play a functional role in the stability and remodeling of the cytoskeleton during fiber cell differentiation in the lens.[32] In cornea, ALDH3 is also suggested to be responsible for absorbing UV-B light.[33]
Co-evolution of lens and cornea through gene sharing
Based on the similarities between lens and cornea, such as abundant water-soluble enzymes, and being derived from ectoderm, the lens and cornea are thought to be co-evolved as a "refraction unit." Gene sharing would maximize light transmission and refraction to the retina by this refraction unit. Studies have shown that many water soluble enzymes/proteins expressed by cornea are identical to taxon-specific lens crystallins, such as ALDH1A1/ η-crystallin, α-enolase/τ-crystallin, and lactic dehydrogenase/ -crystallin. Also, the anuran corneal epithelium, which can transdifferentiate to regenerate the lens, abundantly expresses ubiquitous lens crystallins, α, β and γ, in addition to the taxon-specific crystallin α-enolase/τ-crystallin. Overall, the similarity in expression of these proteins in the cornea and lens, both in abundance and taxon-specificity, supports the idea of co-evolution of lens and cornea through gene sharing.[34]
Relationship to similar concepts
Gene sharing is related to, but distinct from, several concepts in genetics, evolution, and molecular biology. Gene sharing entails multiple effects from the same gene, but unlike pleiotropy, it necessarily involves separate functions at the molecular level. A gene could exhibit pleiotropy when single enzyme function affects multiple phenotypic traits; mutations of a shared gene could potentially affect only a single trait. Gene duplication followed by differential mutation is another phenomenon thought to be a key element in the evolution of protein function, but in gene sharing, there is no divergence of gene sequence when proteins take on new functions; the single polypeptide takes on new roles while retaining old ones. Alternative splicing can result in the production of multiple polypeptides (with multiple functions) from a single gene, but by definition, gene sharing involves multiple functions of a single polypeptide.[5]:8–14
Clinical significance
The multiple roles of moonlighting proteins complicates the determination of phenotype from genotype,[3] hampering the study of inherited metabolic disorders.
The complex phenotypes of several disorders are suspected to be caused by the involvement of moonlighting proteins. The protein GAPDH has at least 11 documented functions, one of which includes apoptosis. Excessive apoptosis is involved in many neurodegenerative diseases, such as Huntington's, Alzheimer's, and Parkinson's as well as in brain ischemia. In one case, GAPDH was found in the degenerated neurons of individuals who had Alzheimer's disease.[3]
Although there is insufficient evidence for definite conclusions, there are well documented examples of moonlighting proteins that play a role in disease. One such disease is tuberculosis. One moonlighting protein in the bacterium M. tuberculosis has a function which counteracts the effects of antibiotics.[9][6] Specifically, M. tuberculosis gains antibiotic resistance against ciprofloxacin from overexpression of Glutamate racemase in vivo.[6]
References
- ^ PDB 3EL3; Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR (December 2009). "Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site". J. Biol. Chem. 284 (52): 36711–36719. doi:10.1074/jbc.M109.064683. PMC 2794785. PMID 19858213. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2794785.
- ^ Jeffery CJ (August 2003). "Moonlighting proteins: old proteins learning new tricks". Trends Genet. 19 (8): 415–7. doi:10.1016/S0168-9525(03)00167-7. PMID 12902157.
- ^ a b c d e f Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM (June 2005). "Single-gene disorders: what role could moonlighting enzymes play?". Am. J. Hum. Genet. 76 (6): 911–924. doi:10.1086/430799. PMC 1196451. PMID 15877277. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1196451.
- ^ a b c Piatigorsky J, Wistow GJ (April 1989). "Enzyme/crystallins: gene sharing as an evolutionary strategy". Cell 57 (2): 197–9. doi:10.1016/0092-8674(89)90956-2. PMID 2649248.
- ^ a b Piatigorsky J (2007). Gene sharing and evolution: the diversity of protein functions. Cambridge: Harvard University Press. ISBN 0-674-02341-2.
- ^ a b c d e Sengupta S, Ghosh S, Nagaraja V (September 2008). "Moonlighting function of glutamate racemase from Mycobacterium tuberculosis: racemization and DNA gyrase inhibition are two independent activities of the enzyme". Microbiology (Reading, Engl.) 154 (Pt 9): 2796–2803. doi:10.1099/mic.0.2008/020933-0. PMID 18757813.
- ^ Piatigorsky J, O'Brien WE, Norman BL, Kalumuck K, Wistow GJ, Borras T, Nickerson JM, Wawrousek EF (May 1988). "Gene sharing by delta-crystallin and argininosuccinate lyase". Proc. Natl. Acad. Sci. U.S.A. 85 (10): 3479–83. doi:10.1073/pnas.85.10.3479. PMC 280235. PMID 3368457. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=280235.
- ^ a b c Jeffery CJ (January 1999). "Moonlighting proteins". Trends Biochem. Sci. 24 (1): 8–11. doi:10.1016/S0968-0004(98)01335-8. PMID 10087914.
- ^ a b c d e f g h i j k Huberts DH, van der Klei IJ (April 2010). "Moonlighting proteins: an intriguing mode of multitasking". Biochim. Biophys. Acta 1803 (4): 520–525. doi:10.1016/j.bbamcr.2010.01.022. PMID 20144902.
- ^ Gancedo C, Flores CL (March 2008). "Moonlighting proteins in yeasts". Microbiol. Mol. Biol. Rev. 72 (1): 197–210. doi:10.1128/MMBR.00036-07. PMC 2268286. PMID 18322039. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2268286.
- ^ Lauble H, Kennedy MC, Beinert H, Stout CD (April 1994). "Crystal structures of aconitase with trans-aconitate and nitrocitrate bound". J. Mol. Biol. 237 (4): 437–451. doi:10.1006/jmbi.1994.1246. PMID 8151704.
- ^ a b c Jeffery CJ (Nov–Dec 2005). "Mass spectrometry and the search for moonlighting proteins". Mass Spectrom. Rev. 24 (6): 772–782. doi:10.1002/mas.20041. PMID 15605385.
- ^ PDB 1P8K; Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL (December 2003). "Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor". Genes Dev. 17 (23): 2875–2888. doi:10.1101/gad.1109003. PMC 289148. PMID 14633971. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=289148.
- ^ PDB 1K87; Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ (February 2003). "Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein". Nat. Struct. Biol. 10 (2): 109–114. doi:10.1038/nsb885. PMID 12514740.
- ^ a b c Jeffery CJ (December 2004). "Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins". Curr. Opin. Struct. Biol. 14 (6): 663–668. doi:10.1016/j.sbi.2004.10.001. PMID 15582389.
- ^ Sriram G, Parr LS, Rahib L, Liao JC, Dipple KM (July 2010). "Moonlighting function of glycerol kinase causes systems-level changes in rat hepatoma cells". Metab. Eng. 12 (4): 332–340. doi:10.1016/j.ymben.2010.04.001. PMC 2949272. PMID 20399282. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2949272.
- ^ Harding JJ, Crabbe MJC (1984). "The lens: development, proteins, metabolism and cataract". In Davson H. The Eye. IB (3 ed.). New York: Academic Press. pp. 207–492.
- ^ a b Wistow GJ, Piatigorsky J (1988). "Lens crystallins: the evolution and expression of proteins for a highly specialized tissue". Annu. Rev. Biochem. 57: 479–504. doi:10.1146/annurev.bi.57.070188.002403. PMID 3052280.
- ^ a b Piatigorsky J (April 1998). "Gene sharing in lens and cornea: facts and implications". Prog Retin Eye Res 17 (2): 145–74. doi:10.1016/S1350-9462(97)00004-9. PMID 9695791.
- ^ Piatigorsky J (2003). "Crystallin genes: specialization by changes in gene regulation may precede gene duplication". J. Struct. Funct. Genomics 3 (1–4): 131–7. doi:10.1023/A:1022626304097. PMID 12836692.
- ^ Ingolia TD, Craig EA (April 1982). "Four small Drosophila heat shock proteins are related to each other and to mammalian alpha-crystallin". Proc. Natl. Acad. Sci. U.S.A. 79 (7): 2360–4. doi:10.1073/pnas.79.7.2360. PMC 346193. PMID 6285380. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=346193.
- ^ de Jong WW, Leunissen JA, Voorter CE (January 1993). "Evolution of the alpha-crystallin/small heat-shock protein family". Mol. Biol. Evol. 10 (1): 103–26. PMID 8450753.
- ^ Aoyama A, Fröhli E, Schäfer R, Klemenz R (March 1993). "Alpha B-crystallin expression in mouse NIH 3T3 fibroblasts: glucocorticoid responsiveness and involvement in thermal protection". Mol. Cell. Biol. 13 (3): 1824–35. PMC 359495. PMID 8441415. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=359495.
- ^ Kegel KB, Iwaki A, Iwaki T, Goldman JE (March 1996). "AlphaB-crystallin protects glial cells from hypertonic stress". Am. J. Physiol. 270 (3 Pt 1): C903–9. PMID 8638673.
- ^ Bhat SP, Nagineni CN (January 1989). "alpha B subunit of lens-specific protein alpha-crystallin is present in other ocular and non-ocular tissues". Biochem. Biophys. Res. Commun. 158 (1): 319–25. doi:10.1016/S0006-291X(89)80215-3. PMID 2912453.
- ^ Kato K, Shinohara H, Kurobe N, Goto S, Inaguma Y, Ohshima K (October 1991). "Immunoreactive alpha A crystallin in rat non-lenticular tissues detected with a sensitive immunoassay method". Biochim. Biophys. Acta 1080 (2): 173–80. doi:10.1016/0167-4838(91)90146-Q. PMID 1932094.
- ^ Dubin RA, Wawrousek EF, Piatigorsky J (March 1989). "Expression of the murine alpha B-crystallin gene is not restricted to the lens". Mol. Cell. Biol. 9 (3): 1083–91. PMC 362698. PMID 2725488. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=362698.
- ^ Holt WS, Kinoshita JH (February 1973). "The soluble proteins of the bovine cornea". Invest Ophthalmol 12 (2): 114–26. PMID 4630510.
- ^ King G, Holmes RS (September 1993). "Human corneal aldehyde dehydrogenase: purification, kinetic characterisation and phenotypic variation". Biochem. Mol. Biol. Int. 31 (1): 49–63. PMID 8260946.
- ^ Wang K, Spector A (May 1994). "The chaperone activity of bovine alpha crystallin. Interaction with other lens crystallins in native and denatured states". J. Biol. Chem. 269 (18): 13601–8. PMID 7909809.
- ^ Blakytny R, Harding JJ (1996). "Prevention of the fructation-induced inactivation of glutathione reductase by bovine alpha-crystallin acting as a molecular chaperone". Ophthalmic Res. 28 Suppl 1: 19–22. PMID 8727959.
- ^ Haynes JI, Duncan MK, Piatigorsky J (September 1996). "Spatial and temporal activity of the alpha B-crystallin/small heat shock protein gene promoter in transgenic mice". Dev. Dyn. 207 (1): 75–88. doi:10.1002/(SICI)1097-0177(199609)207:1<75::AID-AJA8>3.0.CO;2-T. PMID 8875078.
- ^ Algar EM, Abedinia M, VandeBerg JL, Holmes RS (1991). "Purification and properties of baboon corneal aldehyde dehydrogenase: proposed UVR protective role". Adv. Exp. Med. Biol. 284: 53–60. PMID 2053490.
- ^ Jester JV (April 2008). "Corneal crystallins and the development of cellular transparency". Semin. Cell Dev. Biol. 19 (2): 82–93. doi:10.1016/j.semcdb.2007.09.015. PMC 2275913. PMID 17997336. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2275913.
Categories:- Evolutionary biology
- Molecular genetics
- Proteins
- Moonlighting proteins
Wikimedia Foundation. 2010.