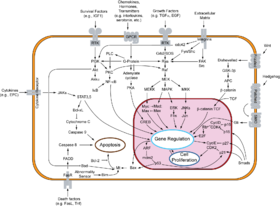
- Systems biology
-
Systems biology is a term used to describe a number of trends in bioscience research, and a movement which draws on those trends. Proponents describe systems biology as a biology-based inter-disciplinary study field that focuses on complex interactions in biological systems, claiming that it uses a new perspective (holism instead of reduction). Particularly from year 2000 onwards, the term is used widely in the biosciences, and in a variety of contexts. An often stated ambition of systems biology is the modeling and discovery of emergent properties, properties of a system whose theoretical description is only possible using techniques which fall under the remit of systems biology. These typically involve cell signaling networks, via long-range allostery[1].
Contents
Overview
Systems biology can be considered from a number of different aspects:
- As a field of study, particularly, the study of the interactions between the components of biological systems, and how these interactions give rise to the function and behavior of that system (for example, the enzymes and metabolites in a metabolic pathway).[2][3]
- As a paradigm, usually defined in antithesis to the so-called reductionist paradigm (biological organisation), although fully consistent with the scientific method. The distinction between the two paradigms is referred to in these quotations:
- "The reductionist approach has successfully identified most of the components and many of the interactions but, unfortunately, offers no convincing concepts or methods to understand how system properties emerge...the pluralism of causes and effects in biological networks is better addressed by observing, through quantitative measures, multiple components simultaneously and by rigorous data integration with mathematical models" Sauer et al[4]
- "Systems biology...is about putting together rather than taking apart, integration rather than reduction. It requires that we develop ways of thinking about integration that are as rigorous as our reductionist programmes, but different....It means changing our philosophy, in the full sense of the term" Denis Noble[5]
- As a series of operational protocols used for performing research, namely a cycle composed of theory, analytic or computational modelling to propose specific testable hypotheses about a biological system, experimental validation, and then using the newly acquired quantitative description of cells or cell processes to refine the computational model or theory.[6] Since the objective is a model of the interactions in a system, the experimental techniques that most suit systems biology are those that are system-wide and attempt to be as complete as possible. Therefore, transcriptomics, metabolomics, proteomics and high-throughput techniques are used to collect quantitative data for the construction and validation of models.
- As the application of dynamical systems theory to molecular biology.
- As a socioscientific phenomenon defined by the strategy of pursuing integration of complex data about the interactions in biological systems from diverse experimental sources using interdisciplinary tools and personnel.
This variety of viewpoints is illustrative of the fact that systems biology refers to a cluster of peripherally overlapping concepts rather than a single well-delineated field. However the term has widespread currency and popularity as of 2007, with chairs and institutes of systems biology proliferating worldwide.
History
Systems biology finds its roots in:[citation needed]
- the quantitative modeling of enzyme kinetics, a discipline that flourished between 1900 and 1970,
- the mathematical modeling of population growth,
- the simulations developed to study neurophysiology, and
- control theory and cybernetics.
One of the theorists who can be seen as one of the precursors of systems biology is Ludwig von Bertalanffy with his general systems theory.[7] One of the first numerical simulations in biology was published in 1952 by the British neurophysiologists and Nobel prize winners Alan Lloyd Hodgkin and Andrew Fielding Huxley, who constructed a mathematical model that explained the action potential propagating along the axon of a neuronal cell.[8] Their model described a cellular function emerging from the interaction between two different molecular components, a potassium and a sodium channel, and can therefore be seen as the beginning of computational systems biology.[9] In 1960, Denis Noble developed the first computer model of the heart pacemaker.[10]
The formal study of systems biology, as a distinct discipline, was launched by systems theorist Mihajlo Mesarovic in 1966 with an international symposium at the Case Institute of Technology in Cleveland, Ohio entitled "Systems Theory and Biology".[11][12]
The 1960s and 1970s saw the development of several approaches to study complex molecular systems, such as the Metabolic Control Analysis and the biochemical systems theory. The successes of molecular biology throughout the 1980s, coupled with a skepticism toward theoretical biology, that then promised more than it achieved, caused the quantitative modelling of biological processes to become a somewhat minor field.[citation needed]
However the birth of functional genomics in the 1990s meant that large quantities of high quality data became available, while the computing power exploded, making more realistic models possible. In 1997, the group of Masaru Tomita published the first quantitative model of the metabolism of a whole (hypothetical) cell.[13]
Around the year 2000, after Institutes of Systems Biology were established in Seattle and Tokyo, systems biology emerged as a movement in its own right, spurred on by the completion of various genome projects, the large increase in data from the omics (e.g. genomics and proteomics) and the accompanying advances in high-throughput experiments and bioinformatics. Since then, various research institutes dedicated to systems biology have been developed. For example, the NIGMS of NIH established a project grant that is currently supporting over ten Systems Biology Centers [4] in the United States. As of summer 2006, due to a shortage of people in systems biology[14] several doctoral training programs in systems biology have been established in many parts of the world. In that same year, the National Science Foundation (NSF) put forward a grand challenge for systems biology in the 21st century to build a mathematical model of the whole cell.[15] In 2011, V. A. Shiva Ayyadurai and C. Forbes Dewey, Jr. of Department of Biological Engineering at the Massachusetts Institute of Technology created CytoSolve, a method to model the whole cell by dynamically integrating multiple molecular pathway models.[16][17]
Associated disciplines
According to the interpretation of Systems Biology as the ability to obtain, integrate and analyze complex data sets from multiple experimental sources using interdisciplinary tools, some typical technology platforms are:
- Phenomics: Organismal variation in phenotype as it changes during its life span.
- Genomics: Organismal deoxyribonucleic acid (DNA) sequence, including intra-organisamal cell specific variation. (i.e. Telomere length variation etc.).
- Epigenomics / Epigenetics: Organismal and corresponding cell specific transcriptomic regulating factors not empirically coded in the genomic sequence. (i.e. DNA methylation, Histone Acetelation etc.).
- Transcriptomics: Organismal, tissue or whole cell gene expression measurements by DNA microarrays or serial analysis of gene expression
- Interferomics: Organismal, tissue, or cell level transcript correcting factors (i.e. RNA interference)
- Translatomics / Proteomics: Organismal, tissue, or cell level measurements of proteins and peptides via two-dimensional gel electrophoresis, mass spectrometry or multi-dimensional protein identification techniques (advanced HPLC systems coupled with mass spectrometry). Sub disciplines include phosphoproteomics, glycoproteomics and other methods to detect chemically modified proteins.
- Metabolomics: Organismal, tissue, or cell level measurements of all small-molecules known as metabolites.
- Glycomics: Organismal, tissue, or cell level measurements of carbohydrates.
- Lipidomics: Organismal, tissue, or cell level measurements of lipids.
In addition to the identification and quantification of the above given molecules further techniques analyze the dynamics and interactions within a cell. This includes:[citation needed]
- Interactomics: Organismal, tissue, or cell level study of interactions between molecules. Currently the authoritative molecular discipline in this field of study is protein-protein interactions (PPI), although the working definition does not pre-clude inclusion of other molecular disciplines such as those defined here.
- NeuroElectroDynamics: Organismal, brain computing function as a dynamic system, underlying biophysical mechanisms and emerging computation by electrical interactions.
- Fluxomics: Organismal, tissue, or cell level measurements of molecular dynamic changes over time.
- Biomics: systems analysis of the biome.
The investigations are frequently combined with large-scale perturbation methods, including gene-based (RNAi, mis-expression of wild type and mutant genes) and chemical approaches using small molecule libraries.[citation needed] Robots and automated sensors enable such large-scale experimentation and data acquisition. These technologies are still emerging and many face problems that the larger the quantity of data produced, the lower the quality.[citation needed] A wide variety of quantitative scientists (computational biologists, statisticians, mathematicians, computer scientists, engineers, and physicists) are working to improve the quality of these approaches and to create, refine, and retest the models to accurately reflect observations.
The systems biology approach often involves the development of mechanistic models, such as the reconstruction of dynamic systems from the quantitative properties of their elementary building blocks.[18][19] For instance, a cellular network can be modelled mathematically using methods coming from chemical kinetics and control theory. Due to the large number of parameters, variables and constraints in cellular networks, numerical and computational techniques are often used.
Other aspects of computer science and informatics are also used in systems biology. These include:
- New forms of computational model, such as the use of process calculi to model biological processes (notable approaches include stochastic π-calculus, BioAmbients, Beta Binders, BioPEPA and Brane calculus) and constraint-based modeling.
- Integration of information from the literature, using techniques of information extraction and text mining.
- Development of online databases and repositories for sharing data and models, approaches to database integration and software interoperability via loose coupling of software, websites and databases, or commercial suits.
- Development of syntactically and semantically sound ways of representing biological models.[citation needed]
See also
- Bioinformatics
- Computational biology
- List of omics topics in biology
- Category:Systems biologists
- List of systems biology research groups
- Network Biology
References
- ^ Bu Z, Callaway DJ (2011). "Proteins MOVE! Protein dynamics and long-range allostery in cell signaling". Advances in Protein Chemistry and Structural Biology. Advances in Protein Chemistry and Structural Biology 83: 163–221. doi:10.1016/B978-0-12-381262-9.00005-7. ISBN 9780123812629. PMID 21570668.
- ^ Snoep, Jacky L; Westerhoff, Hans V (2005). "From isolation to integration, a systems biology approach for building the Silicon Cell". In Alberghina, Lilia; Westerhoff, Hans V. Systems Biology: Definitions and Perspectives. Topics in Current Genetics. 13. Berlin: Springer-Verlag. pp. 13–30. doi:10.1007/b106456. ISBN 978-3-540-22968-1.
- ^ "Systems Biology: the 21st Century Science". Institute for Systems Biology. http://www.systemsbiology.org/Intro_to_Systems_Biology/Systems_Biology_--_the_21st_Century_Science. Retrieved 15 June 2011.
- ^ Sauer, Uwe; Heinemann, Matthias; Zamboni, Nicola (27 April 2007). "GENETICS: Getting Closer to the Whole Picture". Science 316 (5824): 550–551. doi:10.1126/science.1142502. PMID 17463274.
- ^ Noble, Denis (2006). The music of life: Biology beyond the genome. Oxford: Oxford University Press. pp. 176. ISBN 978-0-19-929573-9.
- ^ Kholodenko, Boris N; Sauro, Herbert M (2005). "Mechanistic and modular approaches to modeling and inference of cellular regulatory networks". In Alberghina, Lilia; Westerhoff, Hans V. Systems Biology: Definitions and Perspectives. Topics in Current Genetics. 13. Berlin: Springer-Verlag. pp. 357–451. doi:10.1007/b136809. ISBN 978-3-540-22968-1.
- ^ von Bertalanffy, Ludwig (28 March 1976) [1968]. General System theory: Foundations, Development, Applications. George Braziller. pp. 295. ISBN 9780807604533.
- ^ Hodgkin, Alan L; Huxley, Andrew F (28 August 1952). "A quantitative description of membrane current and its application to conduction and excitation in nerve". Journal of Physiology 117 (4): 500–544. PMC 1392413. PMID 12991237. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1392413. Retrieved 15 June 2011.
- ^ Le Novère, Nicolas (13 June 2007). "The long journey to a Systems Biology of neuronal function". BMC Systems Biology 1: 28. doi:10.1186/1752-0509-1-28. PMC 1904462. PMID 17567903. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1904462. Retrieved 15 June 2011.
- ^ Noble, Denis (5 November 1960). "Cardiac action and pacemaker potentials based on the Hodgkin-Huxley equations". Nature 188 (4749): 495–497. Bibcode 1960Natur.188..495N. doi:10.1038/188495b0. PMID 13729365.
- ^ Mesarovic, Mihajlo D (1968). Systems Theory and Biology. Berlin: Springer-Verlag.
- ^ Rosen, Robert (5 July 1968). "A Means Toward a New Holism". Science 161 (3836): 34–35. doi:10.1126/science.161.3836.34. JSTOR 1724368.
- ^ Tomita, Masaru; Hashimoto, Kenta; Takahashi, Kouichi; Shimizu, Thomas S; Matsuzaki, Yuri; Miyoshi, Fumihiko; Saito, Kanako; Tanida, Sakura et al. (1997). "E-CELL: Software Environment for Whole Cell Simulation". Genome Inform Ser Workshop Genome Inform 8: 147–155. PMID 11072314. http://web.sfc.keio.ac.jp/~mt/mt-lab/publications/Paper/ecell/bioinfo99/btc007_gml.html. Retrieved 15 June 2011.
- ^ Kling, Jim (3 March 2006). "Working the Systems". Science. http://sciencecareers.sciencemag.org/career_magazine/previous_issues/articles/2006_03_03/noDOI.15936087948366349051. Retrieved 15 June 2011.
- ^ The American Association for the Advancement of Science [1]
- ^ National Center for Biotechnology Information [2]
- ^ Massachusetts Institute of Technology [3]
- ^ Gardner, Timothy S; di Bernardo, Diego; Lorenz, David; Collins, James J (4 July 2003). "Inferring Genetic Networks and Identifying Compound Mode of Action via Expression Profiling". Science 301 (5629): 102–105. doi:10.1126/science.1081900. PMID 12843395.
- ^ di Bernardo, Diego; Thompson, Michael J; Gardner, Timothy S; Chobot, Sarah E; Eastwood, Erin L; Wojtovich, Andrew P; Elliott, Sean J; Schaus, Scott E et al. (March 2005). "Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks". Nature Biotechnology 23 (3): 377–383. doi:10.1038/nbt1075. PMID 15765094.
Further reading
- Kitano, Hiroaki (15 October 2001). Foundations of Systems Biology. MIT Press. pp. 320. ISBN 978-0-262-11266-6.
- Werner, Eric (29 March 2007). "All systems go". Nature 446 (7135): 493–494. doi:10.1038/446493a. provides a comparative review of three books:
- Alon, Uri (7 July 2006). An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall. pp. 301. ISBN 978-1-584-88642-6.
- Kaneko, Kunihiko (15 September 2006). Life: An Introduction to Complex Systems Biology. Springer-Verlag. pp. 371. ISBN 978-3-540-32666-3.
- Palsson, Bernhard O (16 January 2006). Systems Biology: Properties of Reconstructed Networks. Cambridge University Press. pp. 334. ISBN 978-0-521-85903-5.
Genomics topics Genome project · Paleopolyploidy · Glycomics · Human Genome Project · Proteomics · Immunomics · Metabolomics · Chemogenomics · Structural genomics · Pharmacogenetics · Pharmacogenomics · Toxicogenomics · Computational genomics · Bioinformatics · Cheminformatics · Systems biologyCategories:
Wikimedia Foundation. 2010.