- Monooxygenase
-
Monooxygenase 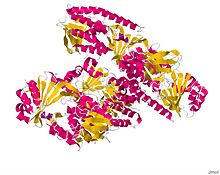
Structure of the Tetx Monooxygenase in complex with the substrate 7-Iodtetracycline.[1] Identifiers Symbol FAD_binding_3 Pfam PF01494 InterPro IPR002938 SCOP 2phh Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Monooxygenases are enzymes that incorporate one hydroxyl group into substrates in many metabolic pathways. In this reaction, two atoms of dioxygen are reduced to one hydroxyl group and one H2O molecule by the concomitant oxidation of NAD(P)H.[2][3]
Contents
Classification
It is classifed as an Oxidoreductase enzyme that catalyzes an electron transfer.
Related Structures
Human proteins containing this domain
COQ6; MICAL1; MICAL2; MICAL2PV1; MICAL2PV2; MICAL3;
References
- ^ PDB 2Y6Q; Volkers G, Palm GJ, Weiss MS, Wright GD, Hinrichs W (April 2011). "Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase". FEBS Lett. 585 (7): 1061–6. doi:10.1016/j.febslet.2011.03.012. PMID 21402075.
- ^ Harayama S, Kok M, Neidle EL (1992). "Functional and evolutionary relationships among diverse oxygenases". Annu. Rev. Microbiol. 46: 565–601. doi:10.1146/annurev.mi.46.100192.003025. PMID 1444267.
- ^ Schreuder HA, van Berkel WJ, Eppink MH, Bunthol C (1999). "Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability". FEBS Lett. 443 (3): 251–255. doi:10.1016/S0014-5793(98)01726-8. PMID 10025942.
This article includes text from the public domain Pfam and InterPro IPR002938
Categories:- Protein domains
- Enzymes
Wikimedia Foundation. 2010.
