- Genotype
-
For a non-technical introduction to the topic, see Introduction to genetics.
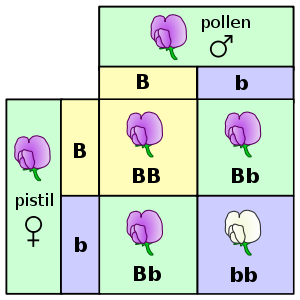 Here the relation between genotype and phenotype is illustrated, using a Punnett square, for the character of petal colour in pea. The letters B and b represent genes for colour and the pictures show the resultant flowers.
Here the relation between genotype and phenotype is illustrated, using a Punnett square, for the character of petal colour in pea. The letters B and b represent genes for colour and the pictures show the resultant flowers.
The genotype is the genetic makeup of a cell, an organism, or an individual (i.e. the specific allele makeup of the individual) usually with reference to a specific character under consideration.[1] For instance, the human CFTR gene, which encodes a protein that transports chloride ions across cell membranes, can be dominant (A) as the normal version of the gene, or recessive (a) as a mutated version of the gene. Individuals receiving two recessive alleles will be diagnosed with Cystic fibrosis. It is generally accepted that inherited genotype, transmitted epigenetic factors, and non-hereditary environmental variation contribute to the phenotype of an individual.
Non-hereditary DNA mutations are not classically understood as representing the individual's genotype. Hence, scientists and physicians sometimes talk for example about the (geno)type of a particular cancer, that is the genotype of the disease as distinct from the diseased.
Contents
Genotype and genomic sequence
Main article: GenomeOne's genotype differs subtly from one's genomic sequence. A sequence is an absolute measure of base composition of an individual, or a representative of a species or group; a genotype typically implies a measurement of how an individual differs or is specialized within a group of individuals or a species. So typically, one refers to an individual's genotype with regard to a particular gene of interest and, in polyploid individuals, it refers to what combination of alleles the individual carries (see homozygous, heterozygous). The genetic constitution of an organism is referred to as its genotype, such as the letters Bb. (B - dominant genotype and b - recessive genotype)
Genotype and Mendelian inheritance
Main article: Mendelian inheritanceThe distinction between genotype and phenotype is commonly experienced when studying family patterns for certain hereditary diseases or conditions, for example, haemophilia. Due to the diploidy of humans (and most animals), there are two alleles for any given gene. These alleles can be the same (homozygous) or different (heterozygous), depending on the individual (see zygote). With a dominant allele, the offspring is guaranteed to inherit the trait in question irrespective of the second allele.
In the case of an albino with a recessive allele (aa), the phenotype depends upon the other allele (Aa, aA or AA). An affected person mating with a heterozygous individual (Aa or aA, also carrier) there is a 50-50 chance the offspring will be albino's phenotype. If a heterozygote mates with another heterozygote, there is 75% chance passing the gene on and only a 25% chance that the gene will be displayed. A homozygous dominant (AA) individual has a normal phenotype and no risk of abnormal offspring. A homozygous recessive individual has an abnormal phenotype and is guaranteed to pass the abnormal gene onto offspring.
In the case of haemophilia, it is sex-linked thus only carried on the X chromosome. Only females can be a carrier in which the abnormality is not displayed. This woman has a normal phenotype, but runs a 50-50 chance, with an unaffected partner, of passing her abnormal gene on to her offspring. If she mated with a man with hemophilia (another carrier) there would be a 75% chance of passing on the gene.
Genotype and mathematics
- Main articles: Genetic programming and evolutionary algorithm
Inspired by the biological concept and usefulness of genotypes, computer science employs simulated phenotypes in genetic programming and evolutionary algorithms. Such techniques can help evolve mathematical solutions to certain types of otherwise difficult problems.
Determining genotype
Main article: GenotypingGenotyping is the process of elucidating the genotype of an individual with a biological assay. Also known as a genotypic assay, techniques include PCR, DNA fragment analysis, allele specific oligonucleotide (ASO) probes, DNA sequencing, and nucleic acid hybridization to DNA microarrays or beads. Several common genotyping techniques include restriction fragment length polymorphism (RFLP), terminal restriction fragment length polymorphism (t-RFLP),[2] amplified fragment length polymorphism (AFLP),[3] and multiplex ligation-dependent probe amplification (MLPA).[4]
DNA fragment analysis can also be used to determine such disease causing genetics aberrations as microsatellite instability (MSI),[5] trisomy[6] or aneuploidy, and loss of heterozygosity (LOH).[7] MSI and LOH in particular have been associated with cancer cell genotypes for colon, breast and cervical cancer.
The most common chromosomal aneuploidy is a trisomy of chromosome 21 which manifests itself as Down syndrome. Current technological limitations typically allow only a fraction of an individual’s genotype to be determined efficiently.
References
- ^ Genotype definition - Medical Dictionary definitions
- ^ Hulce, David; Liu, ChangSheng (Jonathan) (July 2006). "SoftGenetics Application Note - GeneMarker® Software for Terminal-Restriction Fragment Length Polymorphism (T-RFLP) Data Analysis" (PDF). SoftGenetics. http://www.softgenetics.com/T-RFLPapplicationnote.pdf.
- ^ "Keygene.com Homepage"
- ^ "SoftGenetics Application Note - Software for Multiplex Ligation-dependent Probe Amplification (MLPA™)" (PDF). SoftGenetics. April, 2006. http://www.softgenetics.com/MethylationDetectionApplicationNote.pdf. Retrieved 2011-03-13.
- ^ He, Haiguo; Ning, Wan; Liu, Jonathan (March 2007). "SoftGenetics Application Note - Microsatellite Instability Analysis with GeneMarker® Tamela Serensits" (PDF). SoftGenetics. http://www.softgenetics.com/MSIApplicationNote.pdf.
- ^ "SoftGenetics Application Note - GeneMarker® Software for Trisomy Analysis" (PDF). SoftGenetics. November 2006. http://www.softgenetics.com/Trisomyapplicationnote.pdf.
- ^ Serensits, Pamela; He, Haiguo, Ning, Wan; Liu, Jonathan (March 2007). "SoftGenetics Application Note - Loss of Heterozygosity Detection with GeneMarker". SoftGenetics. http://www.softgenetics.com/LOHapplicationnote.pdf.
External links
Categories:
Wikimedia Foundation. 2010.
