- Polyploid
-
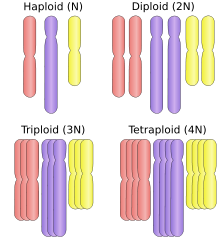
Polyploid is a term used to describe cells and organisms containing more than two paired (homologous) sets of chromosomes. Most eukaryotic species are diploid, meaning they have two sets of chromosomes — one set inherited from each parent. However polyploidy is found in some organisms and is especially common in plants. In addition, polyploidy also occurs in some tissues of animals who are otherwise diploid, such as human muscle tissues.[1] This is known as endopolyploidy. (Monoploid organisms also occur; a monoploid has only one set of chromosomes. These include the vast majority of prokaryotes.)
Polyploidy refers to a numerical change in a whole set of chromosomes. Organisms in which a particular chromosome, or chromosome segment, is under- or overrepresented are said to be aneuploid (from the Greek words meaning "not," "good," and "fold"). Therefore the distinction between aneuploidy and polyploidy is that aneuploidy refers to a numerical change in part of the chromosome set, whereas polyploidy refers to a numerical change in the whole set of chromosomes.[2]
Polyploidy may occur due to abnormal cell division, either during mitosis, or commonly during metaphase I in meiosis.
Polyploidy occurs in some animals, such as goldfish,[3] salmon, and salamanders, but is especially common among ferns and flowering plants (see Hibiscus rosa-sinensis), including both wild and cultivated species. Wheat, for example, after millennia of hybridization and modification by humans, has strains that are diploid (two sets of chromosomes), tetraploid (four sets of chromosomes) with the common name of durum or macaroni wheat, and hexaploid (six sets of chromosomes) with the common name of bread wheat. Many agriculturally important plants of the genus Brassica are also tetraploids. Polyploidization is a mechanism of sympatric speciation because polyploids are usually unable to interbreed with their diploid ancestors.
Polyploidy can be induced in plants and cell cultures by some chemicals: the best known is colchicine, which can result in chromosome doubling, though its use may have other less obvious consequences as well. Oryzalin also will double the existing chromosome content.
Contents
Polyploid types
Polyploid types are labeled according to the number of chromosome sets in the nucleus:
- triploid (three sets; 3x), for example seedless watermelons, common in the phylum Tardigrada[4]
- tetraploid (four sets; 4x), for example Salmonidae fish
- pentaploid (five sets; 5x), for example Kenai Birch (Betula papyrifera var. kenaica)
- hexaploid (six sets; 6x), for example wheat, kiwifruit [5]
- octaploid (eight sets; 8x), for example Acipenser (genus of sturgeon fish), dahlias
- decaploid (ten sets; 10x), for example certain strawberries
- dodecaploid (twelve sets; 12x), for example the plant Celosia argentea and the amphibian Xenopus ruwenzoriensis
Polyploidy in animals
Examples in animals are more common in the 'lower' forms[verification needed] such as flatworms, leeches, and brine shrimp. Polyploid animals are often sterile, so they often reproduce by parthenogenesis. Polyploid lizards are also quite common and parthenogenetic. Polyploid mole salamanders (mostly triploids) are all female and reproduce by kleptogenesis,[6] "stealing" spermatophores from diploid males of related species to trigger egg development but not incorporating the males' DNA into the offspring. While mammalian liver cells are polyploid, rare instances of polyploid mammals are known, but most often result in prenatal death.
An octodontid rodent of Argentina's harsh desert regions, known as the Plains Viscacha-Rat (Tympanoctomys barrerae) has been reported as an exception to this 'rule'. However, careful analysis using chromosome paints shows that there are only two copies of each chromosome in T. barrerae not the four expected if it were truly a tetraploid.[7] The rodent is not a rat, but kin to guinea pigs and chinchillas. Its "new" diploid [2n] number is 102 and so its cells are roughly twice normal size. Its closest living relation is Octomys mimax, the Andean Viscacha-Rat of the same family, whose 2n = 56. It was therefore surmised that an Octomys-like ancestor produced tetraploid (i.e., 4n = 112) offspring that were, by virtue of their doubled chromosomes, reproductively isolated from their parents.
Polyploidy in humans
True polyploidy rarely occurs in humans, although it occurs in some tissues (especially in the liver). Aneuploidy is more common.
Polyploidy occurs in humans in the form of triploidy, with 69 chromosomes (sometimes called 69,XXX), and tetraploidy with 92 chromosomes (sometimes called 92,XXXX). Triploidy, usually due to polyspermy, occurs in about 2–3% of all human pregnancies and ~15% of miscarriages.[citation needed] The vast majority of triploid conceptions end as miscarriage and those that do survive to term typically die shortly after birth. In some cases survival past birth may occur longer if there is mixoploidy with both a diploid and a triploid cell population present.
Triploidy may be the result of either digyny (the extra haploid set is from the mother) or diandry (the extra haploid set is from the father). Diandry is mostly caused by reduplication of the paternal haploid set from a single sperm, but may also be the consequence of dispermic (two sperm) fertilization of the egg.[8] Digyny is most commonly caused by either failure of one meiotic division during oogenesis leading to a diploid oocyte or failure to extrude one polar body from the oocyte. Diandry appears to predominate among early miscarriages while digyny predominates among triploidy that survives into the fetal period.[citation needed] However, among early miscarriages, digyny is also more common in those cases <8.5 weeks gestational age or those in which an embryo is present. There are also two distinct phenotypes in triploid placentas and fetuses that are dependent on the origin of the extra haploid set. In digyny there is typically an asymmetric poorly grown fetus, with marked adrenal hypoplasia and a very small placenta.[citation needed] In diandry, a partial hydatidiform mole develops.[8] These parent-of-origin effects reflect the effects of genomic imprinting.[citation needed]
Complete tetraploidy is more rarely diagnosed than triploidy, but is observed in 1–2% of early miscarriages. However, some tetraploid cells are commonly found in chromosome analysis at prenatal diagnosis and these are generally considered 'harmless'. It is not clear whether these tetraploid cells simply tend to arise during in vitro cell culture or whether they are also present in placental cells in vivo. There are, at any rate, very few clinical reports of fetuses/infants diagnosed with tetraploidy mosaicism.
Mixoploidy is quite commonly observed in human preimplantation embryos and includes haploid/diploid as well as diploid/tetraploid mixed cell populations. It is unknown whether these embryos fail to implant and are therefore rarely detected in ongoing pregnancies or if there is simply a selective process favoring the diploid cells.
Polyploidy in plants
Polyploidy is pervasive in plants and some estimates suggest that 30–80% of living plant species are polyploid, and many lineages show evidence of ancient polyploidy (paleopolyploidy) in their genomes.[9][10][11] Huge explosions in angiosperm species diversity appear to have coincided with the timing of ancient genome duplications shared by many species.[12] It has been established that 15% of angiosperm and 31% of fern speciation events are accompanied by ploidy increase.[13] Polyploid plants can arise spontaneously in nature by several mechanisms, including meiotic or mitotic failures, and fusion of unreduced (2n) gametes.[14] Both autopolyploids (e.g. potato[citation needed]) and allopolyploids (e.g. canola, wheat, cotton) can be found among both wild and domesticated plant species. Most polyploids display heterosis relative to their parental species, and may display novel variation or morphologies that may contribute to the processes of speciation and eco-niche exploitation.[14][10] The mechanisms leading to novel variation in newly formed allopolyploids may include gene dosage effects (resulting from more numerous copies of genome content), the reunion of divergent gene regulatory hierarchies, chromosomal rearrangements, and epigenetic remodeling, all of which affect gene content and/or expression levels.[15][16][17] Many of these rapid changes may contribute to reproductive isolation and speciation.
Lomatia tasmanica is an extremely rare Tasmanian shrub which is triploid and sterile, and reproduction is entirely vegetative with all plants having the same genetic structure.
There are few naturally occurring polyploid conifers. One example is the giant tree Sequoia sempervirens or Coast Redwood which is a hexaploid (6x) with 66 chromosomes (2n = 6x = 66), although the origin is unclear.[18]
Polyploid crops
The induction of polyploidy is a common technique to overcome the sterility of a hybrid species during plant breeding. For example, Triticale is the hybrid of wheat (Triticum turgidum) and rye (Secale cereale). It combines sought-after characteristics of the parents, but the initial hybrids are sterile. After polyploidization, the hybrid becomes fertile and can thus be further propagated to become triticale.
In some situations polyploid crops are preferred because they are sterile. For example many seedless fruit varieties are seedless as a result of polyploidy. Such crops are propagated using asexual techniques such as grafting.
Polyploidy in crop plants is most commonly induced by treating seeds with the chemical colchicine.
Examples of polyploid crops
- Triploid crops: apple, banana, citrus, ginger, watermelon[19]
- Tetraploid crops: apple, durum or macaroni wheat, cotton, potato, cabbage, leek, tobacco, peanut, kinnow, Pelargonium
- Hexaploid crops: chrysanthemum, bread wheat, triticale, oat, kiwifruit[5]
- Octaploid crops: strawberry, dahlia, pansies, sugar cane, oca (Oxalis tuberosa)[20]
Some crops are found in a variety of ploidies: tulips and lilies are commonly found as both diploid and as triploid; daylilies (Hemerocallis cultivars) are available as either diploid or tetraploid; apples and kinnows can be diploid, triploid, or tetraploid.
Terminology
Autopolyploidy
Autopolyploids are polyploids with multiple chromosome sets derived from a single species. Autopolyploids can arise from a spontaneous, naturally occurring genome doubling, like the potato.[citation needed] Others might form following fusion of 2n gametes (unreduced gametes). Bananas and apples can be found as autotriploids. Autopolyploid plants typically display polysomic inheritance, and are therefore often infertile and propagated clonally perfect.
Allopolyploidy
Allopolyploids are polyploids with chromosomes derived from different species. Precisely it is the result of doubling of chromosome number in an F1 hybrid. Triticale is an example of an allopolyploid, having six chromosome sets, allohexaploid, four from wheat (Triticum turgidum) and two from rye (Secale cereale). Amphidiploid is another word for an allopolyploid. Some of the best examples of allopolyploids come from the Brassicas, and the Triangle of U describes the relationships among the three common diploid Brassicas (B. oleracea, B. rapa, and B. nigra) and three allotetraploids (B. napus, B. juncea, and B. carinata) derived from hybridization among the diploids.
Paleopolyploidy
 This phylogenetic tree shows the relationship between the best-documented instances of paleopolyploidy in eukaryotes.
This phylogenetic tree shows the relationship between the best-documented instances of paleopolyploidy in eukaryotes. Main article: Paleopolyploidy
Main article: PaleopolyploidyAncient genome duplications probably occurred in the evolutionary history of all life. Duplication events that occurred long ago in the history of various evolutionary lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as mutations and gene translations gradually make one copy of each chromosome unlike its other copy.
In many cases, these events can be inferred only through comparing sequenced genomes. Examples of unexpected but recently confirmed ancient genome duplications include baker's yeast (Saccharomyces cerevisiae), mustard weed/thale cress (Arabidopsis thaliana), rice (Oryza sativa), and an early evolutionary ancestor of the vertebrates (which includes the human lineage) and another near the origin of the teleost fishes. Angiosperms (flowering plants) have paleopolyploidy in their ancestry. All eukaryotes probably have experienced a polyploidy event at some point in their evolutionary history.
Karyotype
Main article: KaryotypeA karyotype is the characteristic chromosome complement of a eukaryote species.[21][22] The preparation and study of karyotypes is part of cytology and, more specifically, cytogenetics.
Although the replication and transcription of DNA is highly standardized in eukaryotes, the same cannot be said for their karotypes, which are highly variable between species in chromosome number and in detailed organization despite being constructed out of the same macromolecules. In some cases there is even significant variation within species. This variation provides the basis for a range of studies in what might be called evolutionary cytology.
Paralogous
The term is used to describe the relationship among duplicated genes or portions of chromosomes that derived from a common ancestral DNA. Paralogous segments of DNA may arise spontaneously by errors during DNA replication, copy and paste transposons, or whole genome duplications.
Homologous
The term is used to describe the relationship of similar chromosomes that pair at mitosis and meiosis. In a diploid, one homolog is derived from the male parent (sperm) and one is derived from the female parent (egg). During meiosis and gametogenesis, homologous chromosomes pair and exchange genetic material by recombination, leading to the production of sperm or eggs with chromosome haplotypes containing novel genetic variation.
Homoeologous
The term homoeologous, also spelled homeologous, is used to describe the relationship of similar chromosomes or parts of chromosomes brought together following inter-species hybridization and allopolyploidization, and whose relationship was completely homologous in an ancestral species. In allopolyploids, the homologous chromosomes within each parental sub-genome should pair faithfully during meiosis, leading to disomic inheritance; however in some allopolyploids, the homoeologous chromosomes of the parental genomes may be nearly as similar to one another as the homologous chromosomes, leading to tetrasomic inheritance (four chromosomes pairing at meiosis), intergenomic recombination, and reduced fertility.
Example of homoeologous chromosomes
Durum wheat is the result of the inter-species hybridization of two diploid grass species Triticum urartu and Aegilops speltoides. Both the diploid ancestors had two sets of 7 chromosomes, which were similar in terms of size and genes contained on them. Durum wheat contains two sets of chromosomes derived from Triticum urartu and two sets of chromosomes derived from Aegilops speltoides. Each chromosome pair derived from the Triticum urartu parent is homoeologous to the opposite chromosome pair derived from the Aegilops speltoides parent, though each chromosome pair unto itself is homologous.
See also
References
- ^ Parmacek MS, Epstein JA (July 2009). "Cardiomyocyte renewal". N. Engl. J. Med. 361 (1): 86–8. doi:10.1056/NEJMcibr0903347. PMID 19571289.
- ^ Griffiths, Anthony J. F. (1999). An Introduction to genetic analysis. San Francisco: W.H. Freeman. ISBN 0-7167-3520-2.
- ^ Ohno S, Muramoto J, Christian L, Atkin NB (1967). "Diploid-tetraploid relationship among old-world members of the fish family Cyprinidae". Chromosoma 23 (1): 1–9. doi:10.1007/BF00293307. http://www.springerlink.com/content/n3rr5m124g728271/.
- ^ Bertolani R (2001). "Evolution of the reproductive mechanisms in Tardigrades: a review". Zoologischer Anzeiger 240 (3-4): 247–252. doi:10.1078/0044-5231-00032.
- ^ a b The genetic origin of kiwifruit. http://www.actahort.org/books/297/297_5.htm
- ^ Bogart JP, Bi K, Fu J, Noble DW, Niedzwiecki J (February 2007). "Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes". Genome 50 (2): 119–36. doi:10.1139/g06-152. PMID 17546077.
- ^ Svartman M, Stone G, Stanyon R (Aprilt 2005). "Molecular cytogenetics discards polyploidy in mammals". Genomics 85 (4): 425–30. doi:10.1016/j.ygeno.2004.12.004. PMID 15780745.
- ^ a b Baker, Phil; Monga, Ash; Baker, Philip (2006). Gynaecology by ten teachers. London: Arnold. ISBN 0-340-81662-7.
- ^ Meyers LA, Levin DA (June 2006). "On the abundance of polyploids in flowering plants". Evolution 60 (6): 1198–206. doi:10.1111/j.0014-3820.2006.tb01198.x. PMID 16892970.
- ^ a b Rieseberg LH, Willis JH (August 2007). "Plant speciation". Science 317 (5840): 910–4. doi:10.1126/science.1137729. PMC 2442920. PMID 17702935. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2442920.
- ^ Otto SP (November 2007). "The evolutionary consequences of polyploidy". Cell 131 (3): 452–62. doi:10.1016/j.cell.2007.10.022. PMID 17981114.
- ^ de Bodt et al. 2005
- ^ Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (August 2009). "The frequency of polyploid speciation in vascular plants". Proc. Natl. Acad. Sci. U.S.A. 106 (33): 13875–9. doi:10.1073/pnas.0811575106. PMC 2728988. PMID 19667210. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2728988.
- ^ a b Comai L (November 2005). "The advantages and disadvantages of being polyploid". Nat. Rev. Genet. 6 (11): 836–46. doi:10.1038/nrg1711. PMID 16304599.
- ^ Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee HS, Comai L, Madlung A, Doerge RW, Colot V, Martienssen RA (March 2003). "Understanding mechanisms of novel gene expression in polyploids". Trends Genet. 19 (3): 141–7. doi:10.1016/S0168-9525(03)00015-5. PMID 12615008.
- ^ Chen ZJ, Ni Z (March 2006). "Mechanisms of genomic rearrangements and gene expression changes in plant polyploids". Bioessays 28 (3): 240–52. doi:10.1002/bies.20374. PMC 1986666. PMID 16479580. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1986666.
- ^ Chen ZJ (2007). "Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids". Annu Rev Plant Biol 58: 377–406. doi:10.1146/annurev.arplant.58.032806.103835. PMC 1949485. PMID 17280525. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1949485.
- ^ Ahuja MR, Neale DB (2002). "Origins of Polyploidy in Coast Redwood (Sequoia sempervirens (D. DON) ENDL.) and Relationship of Coast Redwood to other Genera of Taxodiaceae". Silvae Genetica 51: 2–3.
- ^ Seedless Fruits Make Others Needless
- ^ Emshwiller, E. (2006). "Origins of polyploid crops: The example of the octaploid tuber crop Oxalis tuberosa". In M.A. Zeder; D. Decker-Walters; E. Emshwiller; D. Bradley; B.D. Smith. Documenting domestication: new genetic and archaeological paradigms. University of California Press. pp. 153–168.
- ^ White M.J.D. 1973. The chromosomes. 6th ed, Chapman & Hall, London. p28
- ^ Stebbins G.L. 1950. Variation and evolution in plants. Chapter XII: The Karyotype. Columbia University Press N.Y.
Further reading
- Snustad, P. et al. 2006. Principles of Genetics, 4th ed. John Wiley & Sons, Inc. Hoboken, NJ ISBN 100-471-69939-X
- Arabidopsis Genome Initiative (December 2000). "Analysis of the genome sequence of the flowering plant Arabidopsis thaliana". Nature 408 (6814): 796–815. doi:10.1038/35048692. PMID 11130711.
- Eakin GS, Behringer RR (December 2003). "Tetraploid development in the mouse". Dev. Dyn. 228 (4): 751–66. doi:10.1002/dvdy.10363. PMID 14648853.
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC (November 2007). "Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype". Plant Cell 19 (11): 3403–17. doi:10.1105/tpc.107.054346. PMC 2174891. PMID 18024568. http://www.plantcell.org/cgi/pmidlookup?view=long&pmid=18024568.
- Gregory, T.R. & Mable, B.K. (2005). Polyploidy in animals. In The Evolution of the Genome (edited by T.R. Gregory). Elsevier, San Diego, pp. 427–517.
- Jaillon O, Aury JM, Brunet F, et al. (October 2004). "Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype". Nature 431 (7011): 946–57. doi:10.1038/nature03025. PMID 15496914.
- Paterson AH, Bowers JE, Van de Peer Y, Vandepoele K (March 2005). "Ancient duplication of cereal genomes". New Phytol. 165 (3): 658–61. doi:10.1111/j.1469-8137.2005.01347.x. PMID 15720677.
- Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y (2003). "Investigating ancient duplication events in the Arabidopsis genome". J. Struct. Funct. Genomics 3 (1-4): 117–29. doi:10.1023/A:1022666020026. PMID 12836691. http://www.kluweronline.com/art.pdf?issn=1345-711X&volume=3&page=117.
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (October 2002). "The hidden duplication past of Arabidopsis thaliana". Proc. Natl. Acad. Sci. U.S.A. 99 (21): 13627–32. doi:10.1073/pnas.212522399. PMC 129725. PMID 12374856. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=12374856.
- Soltis DE, Soltis PS, Schemske DW, Hancock JF, Thompson JN, Husband BC, Judd WS (2007). "Autopolyploidy in angiosperms: have we grossly underestimated the number of species?". Taxon 56 (1): 13–30.
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS (2010). "What we still don't know about polyploidy". Taxon 59: 1387–1403.
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y (March 2003). "Genome duplication, a trait shared by 22000 species of ray-finned fish". Genome Res. 13 (3): 382–90. doi:10.1101/gr.640303. PMC 430266. PMID 12618368. http://www.genome.org/cgi/pmidlookup?view=long&pmid=12618368.
- Tate, J.A., Soltis, D.E., & Soltis, P.S. (2005). Polyploidy in plants. In The Evolution of the Genome (edited by T.R. Gregory). Elsevier, San Diego, pp. 371–426.
- Van de Peer Y, Taylor JS, Meyer A (2003). "Are all fishes ancient polyploids?". J. Struct. Funct. Genomics 3 (1-4): 65–73. doi:10.1023/A:1022652814749. PMID 12836686. http://www.kluweronline.com/art.pdf?issn=1345-711X&volume=3&page=65.
- Van de Peer Y (2004). "Tetraodon genome confirms Takifugu findings: most fish are ancient polyploids". Genome Biol. 5 (12): 250. doi:10.1186/gb-2004-5-12-250. PMC 545788. PMID 15575976. http://genomebiology.com/content/5/12/250.
- Van de Peer, Y. and Meyer, A. (2005). Large-scale gene and ancient genome duplications. In The Evolution of the Genome (edited by T.R. Gregory). Elsevier, San Diego, pp. 329–368
- Wolfe KH, Shields DC (June 1997). "Molecular evidence for an ancient duplication of the entire yeast genome". Nature 387 (6634): 708–13. doi:10.1038/42711. PMID 9192896.
- Wolfe KH (May 2001). "Yesterday's polyploids and the mystery of diploidization". Nat. Rev. Genet. 2 (5): 333–41. doi:10.1038/35072009. PMID 11331899.
External links
- Polyploidy on Kimball's Biology Pages
- The polyploidy portal a community-editable project with information, research, education, and a bibliography about polyploidy.
Genetics: chromosomes General Classification Evolution Structure Speciation Basic concepts Modes of speciation Auxiliary mechanisms Intermediate stages Categories:
Wikimedia Foundation. 2010.