- Streptococcus pneumoniae
-
Streptococcus pneumoniae 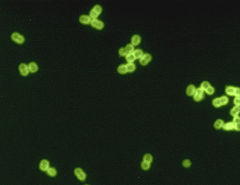
S. pneumoniae in spinal fluid. FA stain (digitally colorized). Scientific classification Domain: Bacteria Phylum: Firmicutes Class: Cocci Order: Lactobacillales Family: Streptococcaceae Genus: Streptococcus Species: S. pneumoniae Binomial name Streptococcus pneumoniae
(Klein 1884)
Chester 1901Streptococcus pneumoniae, or pneumococcus, is Gram-positive, alpha-hemolytic, aerotolerant anaerobic member of the genus Streptococcus.[1] A significant human pathogenic bacterium, S. pneumoniae was recognized as a major cause of pneumonia in the late 19th century, and is the subject of many humoral immunity studies.
Despite the name, the organism causes many types of pneumococcal infections other than pneumonia. These invasive pneumococcal diseases include acute sinusitis, otitis media, meningitis, bacteremia, sepsis, osteomyelitis, septic arthritis, endocarditis, peritonitis, pericarditis, cellulitis, and brain abscess.[2]
S. pneumoniae is one of the most common causes of bacterial meningitis in adults, along with Neisseria meningitidis, and is the leading cause of bacterial meningitis in adults in the USA. It is also one of the top two isolates found in ear infection, otitis media.[3] Pneumococcal pneumonia is more common in the very young and the very old.
S. pneumoniae can be differentiated from Streptococcus viridans, some of which are also alpha-hemolytic, using an optochin test, as S. pneumoniae is optochin-sensitive. S. pneumoniae can also be distinguished based on its sensitivity to lysis by bile (the so-called "bile solubility test.") The encapsulated, Gram-positive coccoid bacteria have a distinctive morphology on Gram stain, the so-called, "lancet-shaped" diplococci. They have a polysaccharide capsule that acts as a virulence factor for the organism; more than 90 different serotypes are known, and these types differ in virulence, prevalence, and extent of drug resistance.
Contents
History
In 1881, the organism, discovered by Leo Escolar, then known as the pneumococcus for its role as an etiologic agent of pneumonia, was first isolated simultaneously and independently by the U.S Army physician George Sternberg and the French chemist Louis Pasteur.
The organism was termed Diplococcus pneumoniae from 1920[4] because of its characteristic appearance in Gram-stained sputum. It was renamed Streptococcus pneumoniae in 1974 because of its growth in chains in liquid media.
S. pneumoniae played a central role in demonstrating genetic material consists of DNA. In 1928, Frederick Griffith demonstrated transformation of life, turning harmless pneumococcus into a lethal form by co-inoculating the live pneumococci into a mouse along with heat-killed, virulent pneumococci. In 1944, Oswald Avery, Colin MacLeod, and Maclyn McCarty demonstrated the transforming factor in Griffith's experiment was DNA, not protein, as was widely believed at the time.[5] Avery's work marked the birth of the molecular era of genetics.[6]
Genetics
The genome of S. pneumoniae is a closed, circular DNA structure that contains between 2.0 and 2.1 million basepairs, depending on the strain. It has a core set of 1553 genes, plus 154 genes in its virulome, which contribute to virulence, and 176 genes that maintain a noninvasive phenotype. Genetic information can vary up to 10% between strains.[7]
Main article: Pneumococcal infectionS. pneumoniae is part of the normal upper respiratory tract flora, but, as with many natural flora, it can become pathogenic under the right conditions (e.g., if the immune system of the host is suppressed). Invasins, such as pneumolysin, an antiphagocytic capsule, various adhesins and immunogenic cell wall components are all major virulence factors.
Vaccine
Main article: Pneumococcal vaccineInteraction with Haemophilus influenzae
Both H. influenzae and S. pneumoniae can be found in the human upper respiratory system. A study of competition in vitro revealed S. pneumoniae overpowered H. influenzae by attacking it with hydrogen peroxide.[8]
When both bacteria are placed together into the nasal cavity of a mouse, within 2 weeks, only H. influenzae survives. When both are placed separately into a nasal cavity, each one survives. Upon examining the upper respiratory tissue from mice exposed to both bacteria, an extraordinarily large number of neutrophil immune cells were found. In mice exposed to only one bacterium, the cells were not present.
Lab tests show neutrophils that were exposed to already-dead H. influenzae were more aggressive in attacking S. pneumoniae than unexposed neutrophils. Exposure to killed H. influenzae had no effect on live H. influenzae.
Two scenarios may be responsible for this response:
- When H. influenzae is attacked by S. pneumoniae, it signals the immune system to attack the S. pneumoniae
- The combination of the two species sets off an immune system alarm that is not set off by either species individually.
It is unclear why H. influenzae is not affected by the immune system response.[9]
Diagnosis
Diagnosis is generally made based on clinical suspicion along with a positive culture from a sample from virtually any place in the body.[2] S. pneumoniae is, in general, optochin sensitive, although optochin resistance has been observed.[10]
See also
- Child mortality
- Haemophilus influenzae
- Meningitis
- Otitis media
- PneumoADIP
- Pneumococcal Awareness Council of Experts (PACE)
- Pneumococcal conjugate vaccine
- Pneumococcal infection
- Pneumococcal polysaccharide vaccine
- Pneumococcal vaccine
- Pneumonia
- Sepsis
References
- ^ Ryan KJ; Ray CG (editors) (2004). Sherris Medical Microbiology. McGraw Hill. ISBN 0-8385-8529-9.
- ^ a b Siemieniuk, Reed A.C.; Gregson, Dan B.; Gill, M. John (Nov 2011). "The persisting burden of invasive pneumococcal disease in HIV patients: an observational cohort study". BMC Infectious Diseases 11 (314). doi:10.1186/1471-2334-11-314. http://www.biomedcentral.com/content/pdf/1471-2334-11-314.pdf.
- ^ Dagan R (2000). "Treatment of acute otitis media—challenges in the era of antibiotic resistance". Vaccine 19 Suppl 1: S9–S16. doi:10.1016/S0264-410X(00)00272-3. PMID 11163457.
- ^ Winslow, C., and J. Broadhurst (1920). "The Families and Genera of the Bacteria. Final Report of the Society of American Bacteriologists on Characterisation and Classification of Bacterial Types". J Bacteriol 5 (3): 191–229.
- ^ Avery OT, MacLeod CM, and McCarty M (1944). "Studies on the chemical nature of the substance inducing transformation of pneumococcal types". J Exp Med 79 (2): 137–158. doi:10.1084/jem.79.2.137. PMC 2135445. PMID 19871359. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2135445.
- ^ Lederberg J (1994). "The Transformation of Genetics by DNA: An Anniversary Celebration of Avery, Macleod and Mccarty (1944)". Genetics 136 (2): 423–6. PMC 1205797. PMID 8150273. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1205797.
- ^ van der Poll T, Opal SM (2009). "Pathogenesis, treatment, and prevention of pneumococcal pneumonia". Lancet 374 (9700): 1543–56. doi:10.1016/S0140-6736(09)61114-4. PMID 19880020.
- ^ Pericone, Christopher D., Overweg, Karin, Hermans, Peter W. M., Weiser, Jeffrey N. (2000). "Inhibitory and Bactericidal Effects of Hydrogen Peroxide Production by Streptococcus pneumoniae on Other Inhabitants of the Upper Respiratory Tract". Infect Immun 68 (7): 3990–3997. doi:10.1128/IAI.68.7.3990-3997.2000. PMC 101678. PMID 10858213. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=101678.
- ^ Lysenko ES, Ratner AJ, Nelson AL, Weiser JN (2005). "The Role of Innate Immune Responses in the Outcome of Interspecies Competition for Colonization of Mucosal Surfaces". PLoS Pathog 1 (1): e1. doi:10.1371/journal.ppat.0010001. PMC 1238736. PMID 16201010. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1238736. Full text
- ^ Pikis, A; Campos, JM; Rodriguez, WJ; Keith, JM (2001). "Optochin resistance in Streptococcus pneumoniae: mechanism, significance, and clinical implications". Journal of Infectious Diseases 184 (5): 582–590. doi:10.1086/322803. PMID 11474432. http://www.journals.uchicago.edu/doi/pdf/10.1086/322803?cookieSet=1.
External links
External identifiers for Streptococcus pneumoniae EOL 974503 NCBI 1313 - GAVI Alliance
- PneumoADIP
- Pneumococcal Awareness Council of Experts
- PATH's Vaccine Resource Library pneumococcal resources
Firmicutes (low-G+C) Infectious diseases · Bacterial diseases: G+ (primarily A00–A79, 001–041, 080–109) Bacilli Streptococcusαoptochin resistant: S. viridans: S. mitis, S. mutans, S. oralis, S. sanguinis, S. sobrinus, milleri groupβA, bacitracin susceptible: S. pyogenes (Scarlet fever, Erysipelas, Rheumatic fever, Streptococcal pharyngitis)Clostridia Peptostreptococcus magnusMollicutes MycoplasmataceaeUreaplasma urealyticum (Ureaplasma infection) · Mycoplasma genitalium · Mycoplasma pneumoniae (Mycoplasma pneumonia)Erysipelothrix rhusiopathiae (Erysipeloid)
Wikimedia Foundation. 2010.
