- mir-2 microRNA precursor
-
mir-2 microRNA precursor 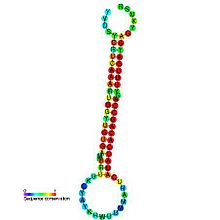
Predicted secondary structure and sequence conservation of mir-2 Identifiers Symbol mir-2 Rfam RF00047 miRBase MI0000117 miRBase family MIPF0000049 Other data RNA type Gene; miRNA Domain(s) Eukaryota GO 0035195 0035068 SO 0001244 This family represents the microRNA (miRNA) precursors mir-2 and mir-13 (MIPF0000049). Genomic loci encoding miR-2 products have been described in a wide range of species and these products have been experimentally verified in multiple invertebrates. The miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 3' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference. MicroRNAs are numbered based on the mature sequence. This family also contains the mir-13 precursor which, despite giving rise to a distinct mature sequence appears to be related in sequence.
By specific antisense-mediated inactivation of miR-2 products, Boutla et al. first demonstrated a fundamental role of mir-2 family during development.[1] Leaman et al.. showed that the miR-2 family regulates cell survival by translational repression of proapoptotic factors [2]
References
- ^ Boutla, A; Delidakis, C; Tabler, M (2003). "Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes". Nucleic Acids Research 31 (17): 4973-4980. doi:10.1093/nar/gkg707. PMID 12930946.
- ^ Leaman, D; Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U (2005). "Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development". Cell 121 (7): 1097–1108. doi:10.1016/j.cell.2005.04.016. PMID 15989958.
External links
miRNA precursor families 1-100 101-200 201+ Other Categories:- MicroRNA
- Molecular and cellular biology stubs
Wikimedia Foundation. 2010.
