- mir-395 microRNA precursor family
-
mir-395 microRNA precursor family 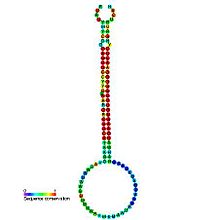
Predicted secondary structure and sequence conservation of mir-395 Identifiers Symbol mir-395 Rfam RF00451 miRBase MI0001007 miRBase family MIPF0000016 Other data RNA type Gene; miRNA Domain(s) Eukaryota GO 0035195 0035068 SO 0001244 mir-395 is a non-coding RNA called a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-395 is thought to target mRNAs coding for ATP sulphurylases.[1] The mature sequence is excised from the 3' arm of the hairpin.
miR-395 is upregulated in Arabidopsis during sulphate-limited conditions, when the mature miRNA then regulates sulphur transporters and ATP sulphurylases.[2][3]
References
- ^ Jones-Rhoades, MW; Bartel DP (2004). "Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA". Mol Cell 14 (6): 787–799. doi:10.1016/j.molcel.2004.05.027. PMID 15200956.
- ^ Liang, G; Yang, F, Yu, D (2010 Jun 1). "MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana.". The Plant journal : for cell and molecular biology 62 (6): 1046–57. PMID 20374528.
- ^ Liang, G; Yu, D (2010 Oct). "Reciprocal regulation among miR395, APS and SULTR2;1 in Arabidopsis thaliana.". Plant signaling & behavior 5 (10): 1257–9. PMID 20935495.
External links
miRNA precursor families 1-100 101-200 201+ Other Categories:- MicroRNA
- Molecular and cellular biology stubs
Wikimedia Foundation. 2010.
