- mir-172 microRNA precursor family
-
mir-172 microRNA precursor family 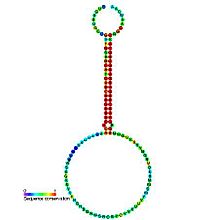
Predicted secondary structure and sequence conservation of mir-172 Identifiers Symbol mir-172 Rfam RF00452 miRBase MI0000215 miRBase family MIPF0000035 Other data RNA type Gene; miRNA Domain(s) Eukaryota GO 0035195 0035068 SO 0001244 The mir-172 microRNA is thought to target mRNAs coding for APETALA2-like transcription factors.[1] It has been verified experimentally in the model plant, Arabidopsis thaliana (mouse-ear cress).[2][3] The mature sequence is excised from the 3' arm of the hairpin.
References
- ^ Jones-Rhoades, MW; Bartel DP (2004). "Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA". Mol Cell 14 (6): 787–799. doi:10.1016/j.molcel.2004.05.027. PMID 15200956.
- ^ Park, W; Li J, Song R, Messing J, Chen X (2002). "CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana". Curr Biol 12 (17): 1484–1495. doi:10.1016/S0960-9822(02)01017-5. PMID 12225663.
- ^ Mette, MF; van der Winden J, Matzke M, Matzke AJ (2002). "Short RNAs can identify new candidate transposable element families in Arabidopsis". Plant Physiol 130 (1): 6–9. doi:10.1104/pp.007047. PMC 1540252. PMID 12226481. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1540252.
Further reading
- Zhang B, Pan X (July 2009). "Expression of microRNAs in cotton". Mol. Biotechnol. 42 (3): 269–74. doi:10.1007/s12033-009-9163-y. PMID 19288227.
- Abdurakhmonov IY, Devor EJ, Buriev ZT, et al. (2008). "Small RNA regulation of ovule development in the cotton plant, G. hirsutum L". BMC Plant Biol. 8: 93. doi:10.1186/1471-2229-8-93. PMC 2564936. PMID 18793449. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2564936.
External links
miRNA precursor families 1-100 101-200 201+ Other Categories:- Molecular and cellular biology stubs
- MicroRNA
Wikimedia Foundation. 2010.
