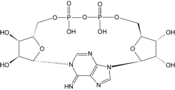
- Cyclic ADP-ribose
-
Cyclic ADP-ribose Identifiers CAS number 119340-53-3 PubChem 123847 MeSH Cyclic+ADP-Ribose IUPHAR ligand 2445 Properties Molecular formula C15H21N5O13P2 Molar mass 541.301  (verify) (what is:
(verify) (what is:  /
/ ?)
?)
Except where noted otherwise, data are given for materials in their standard state (at 25 °C, 100 kPa)Infobox references Cyclic ADP Ribose, frequently abbreviated as cADPR, is a cyclic adenine nucleotide (like cAMP) with two phosphate groups present on 5' OH of the adenosine (like ADP), further connected to another ribose at the 5' position, which, in turn, closes the cycle by glycosidic bonding to the nitrogen 1 (N1) of the same adenine base (whose position N9 has the glycosidic bond to the other ribose)[1][2]. The N1-glycosidic bond to adenine is what distinguishes cADPR from ADP-ribose (ADPR), the non-cyclic analog. cADPR is produced from nicotinamide adenine dinucleotide (NAD+) by ADP-ribosyl cyclases (EC 3.2.2.5) as part of a second messenger system.
Contents
Function
cADPR is a cellular messenger for calcium signaling.[3] It is the physiological allosteric modulator of the ryanodine receptor (RyR), which stimulates calcium-induced calcium release at lower cytosolic concentrations of Ca2+. RyR activation with high concentration of caffeine is partly due to caffeine's mimicking the binding of cADPR to RyRs. Whether the action is by direct binding to RyR or indirect (through binding with FKBP12.6) is debated. Some reports suggest that cADPR binding makes FKBP12.6, which normally binds RyR2, to fall off the RYR2.
Metabolism
cADPR and ADPR are synthesized from NAD+ by the bifunctional ectoenzymes of the CD38 family (also includes the GPI-anchored CD157 and the specific, monofunctional ADP ribosyl cyclase of the mollusc Aplysia).[4][5][6] The same enzymes are also capable of hydrolyzing cADPR to ADPR. Catalysis proceeds via a covalently bound intermediate. The hydrolysis reaction is inhibited by ATP, and cADPR may accumulate. Synthesis and degradation of cADPR by enzymes of the CD38 family involve, respectively, the formation and the hydrolysis of the N1-glycosidic bond. In 2009, the first enzyme able to hydrolyze the phosphoanhydride linkage of cADPR, i.e. the one between the two phosphate groups, has been reported.[7]
See also
- NAADP
- IP3
- ADP-ribose
References
- ^ Lee HC, Walseth TF, Bratt GT, Hayes RN, Clapper DL (1989). "Structural determination of a cyclic metabolite of NAD+ with intracellular Ca2+-mobilizing activity". J. Biol. Chem. 264 (3): 1608–15. PMID 2912976.
- ^ Lee HC, Aarhus R, Levitt D (1994). "The crystal structure of cyclic ADP-ribose". Nat. Struct. Biol. 1 (3): 143–4. doi:10.1038/nsb0394-143. PMID 7656029.
- ^ Guse AH (2004). "Regulation of calcium signaling by the second messenger cyclic adenosine diphosphoribose (cADPR)". Curr. Mol. Med. 4 (3): 239–48. doi:10.2174/1566524043360771. PMID 15101682.
- ^ Prasad GS, McRee DE, Stura EA, Levitt DG, Lee HC, Stout CD (1996). "Crystal structure of Aplysia ADP-ribosyl cyclase, a homolog of the bifunctional ectozyme CD38". Nat. Struct. Biol. 3 (11): 957–64. doi:10.1038/nsb1196-957. PMID 8901875.
- ^ Liu Q, Kriksunov IA, Graeff R, Munshi C, Lee HC, Hao Q (2005). "Crystal structure of the human CD38 extracellular domain". Structure 13 (9): 1331–9. doi:10.1016/j.str.2005.05.012. PMID 16154090.
- ^ Guse AH (2004). "Biochemistry, biology, and pharmacology of cyclic adenosine diphosphoribose (cADPR)". Curr. Med. Chem. 11 (7): 847–55. doi:10.2174/0929867043455602. PMID 15078169.
- ^ Canales J, Fernández A, Rodrigues JR, Ferreira R, Ribeiro JM, Cabezas A, Costas MJ, Cameselle JC (2009). "Hydrolysis of the phosphoanhydride linkage of cyclic ADP-ribose by the Mn2+-dependent ADP-ribose/CDP-alcohol pyrophosphatase". FEBS Lett. 583 (10): 1593–8. doi:10.1016/j.febslet.2009.04.023. PMID 19379742.
External links
The web page of Dr. Hon Cheung Lee, the discoverer of cyclic ADP-ribose.
Cyclic ADP-ribose and NAADP. The first book on these two second messengers.
Cell signaling: calcium signaling / calcium metabolism Cell membrane Ion pumpsAdhesion moleculesOtherIntracellular signaling
& calc. regulationSecond messengersChelators and calcium sensorsCalbindin · S100 · pervalbumin · Calretinin · Calsequestrin · Sarcalumenin · Phospholamban · SynaptotagminsCytoskeleton remodeling proteinsOtherCalcium-binding
protein domainsExtracellular ligands Calcium-binding proteins B trdu: iter (nrpl/grfl/cytl/horl), csrc (lgic, enzr, gprc, igsr, intg, nrpr/grfr/cytr), itra (adap, gbpr, mapk), calc, lipd; path (hedp, wntp, tgfp+mapp, notp, jakp, fsap, hipp, tlrp) Nucleic acid constituents Nucleobase Nucleoside Nucleotide
(Nucleoside monophosphate)Nucleoside diphosphate Nucleoside triphosphate biochemical families: prot · nucl · carb (glpr, alco, glys) · lipd (fata/i, phld, strd, gllp, eico) · amac/i · ncbs/i · ttpy/i Categories:- Nucleotides
Wikimedia Foundation. 2010.