- Cyclophilin
-
peptidylprolyl isomerase A (cyclophilin A) 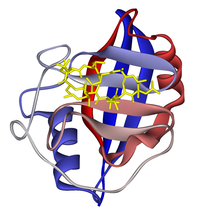
Ribbon diagram of cyclophilin A in complex with ciclosporin (yellow). From PDB 1CWA. Identifiers Symbol PPIA Entrez 5478 HUGO 9253 OMIM 123840 RefSeq NM_203430 UniProt Q3KQW3 Other data EC number 5.2.1.8 Locus Chr. 7 p13 Pro_isomerase 
x-ray structure of peptidyl-prolyl cis-trans isomerase a, ppia, rv0009, from mycobacterium tuberculosis. Identifiers Symbol Pro_isomerase Pfam PF00160 Pfam clan CL0475 InterPro IPR002130 PROSITE PDOC00154 SCOP 1cyh Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Cyclophilins are a family of proteins from vertebrates and other organisms that bind to cyclosporine, an immunosuppressant which is usually used to suppress rejection after internal organ transplants [1]. These proteins have peptidyl prolyl isomerase activity, which catalyzes the isomerization of peptide bonds from trans form to cis form at proline residues and facilitates protein folding.
Cyclophilin A is a cytosolic and highly abundant protein. The protein belongs to a family of isozymes, including cyclophilins B and C, and natural killer cell cyclophilin-related protein.[2][3][4] Major isoforms have been found throughout the cell, including the ER, and some are even secreted.
Contents
Cyclophilin A (CycA)
Cyclophilin A also known as peptidylprolyl isomerase A, which is found in the cytosol, has a beta barrel structure with two alpha helices and a beta-sheet. Other cyclophilins have similar structures to cyclophilin A. The ciclosporin-cyclophilin A complex inhibits a calcium/calmodulin-dependent phosphatase, calcineurin, the inhibition of which is thought to suppress organ rejection by halting the production of the pro-inflammatory molecules TNF alpha and interleukin 2.
Cyclophilin A is also known to be recruited by the Gag polyprotein during HIV-1 virus infection, and its incorporation into new virus particles is essential for HIV-1 infectivity.
Cyclophilin D
Cyclophilin D, which is located in the matrix of mitochondria, is a component of the mitochondrial permeability transition pore. The pore opening raises the permeability of the mitochondrial inner membrane, allows influx of cytosolic molecules into the mitochondrial matrix, increases the matrix volume, and disrupts the mitochondrial outer membrane. As a result, the mitochondria fall into a functional disorder, so the opening of the pore plays an important role in cell death. Cyclophilin D is thought to regulate the opening of the pore because cyclosporin A (CsA), which binds to CyP-D, inhibits the pore opening.
Examples
Human genes encoding proteins containing the cyclophilin type peptidyl-prolyl cis-trans isomerase domain include:
References
- ^ Stamnes MA, Rutherford SL, Zuker CS (September 1992). "Cyclophilins: a new family of proteins involved in intracellular folding". Trends Cell Biol. 2 (9): 272–6. doi:10.1016/0962-8924(92)90200-7. PMID 14731520.
- ^ Trandinh CC, Pao GM, Saier MH (December 1992). "Structural and evolutionary relationships among the immunophilins: two ubiquitous families of peptidyl-prolyl cis-trans isomerases". FASEB J. 6 (15): 3410–20. PMID 1464374.
- ^ Galat A (September 1993). "Peptidylproline cis-trans-isomerases: immunophilins". Eur. J. Biochem. 216 (3): 689–707. doi:10.1111/j.1432-1033.1993.tb18189.x. PMID 8404888.
- ^ Hacker J, Fischer G (November 1993). "Immunophilins: structure-function relationship and possible role in microbial pathogenicity". Mol. Microbiol. 10 (3): 445–56. doi:10.1111/j.1365-2958.1993.tb00917.x. PMID 7526121.
External links
Isomerases: geometric (EC 5.2) 5.2.1 B enzm: 1.1/2/3/4/5/6/7/8/10/11/13/14/15-18, 2.1/2/3/4/5/6/7/8, 2.7.10, 2.7.11-12, 3.1/2/3/4/5/6/7, 3.1.3.48, 3.4.21/22/23/24, 4.1/2/3/4/5/6, 5.1/2/3/4/99, 6.1-3/4/5-6 Transmembrane receptors: Immunoglobulin superfamily immune receptors Antibody receptor:
Fc receptorSecretoryAntigen receptor Antigen receptorAccessory moleculesT cellsAntigen receptorAccessory moleculesCytokine receptor see cytokine receptorsKiller-cell IG-like receptors Leukocyte IG-like receptors B trdu: iter (nrpl/grfl/cytl/horl), csrc (lgic, enzr, gprc, igsr, intg, nrpr/grfr/cytr), itra (adap, gbpr, mapk), calc, lipd; path (hedp, wntp, tgfp+mapp, notp, jakp, fsap, hipp, tlrp) Categories:- Genes on chromosome 7
- EC 5.2.1
- Isomerases
Wikimedia Foundation. 2010.
