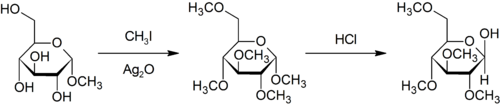- Methylation
-
In the chemical sciences, methylation denotes the addition of a methyl group to a substrate or the substitution of an atom or group by a methyl group. Methylation is a form of alkylation with, to be specific, a methyl group, rather than a larger carbon chain, replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences.
In biological systems, methylation is catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA metabolism. Methylation of heavy metals can also occur outside of biological systems. Chemical methylation of tissue samples is also one method for reducing certain histological staining artifacts.
Contents
In biology
Epigenetics
Methylation contributing to epigenetic inheritance can occur through either DNA methylation or protein methylation.
DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sites, that is, where a cytosine is directly followed by a guanine in the DNA sequence). This methylation results in the conversion of the cytosine to 5-methylcytosine. The formation of Me-CpG is catalyzed by the enzyme DNA methyltransferase. Human DNA has about 80%-90% of CpG sites methylated, but there are certain areas, known as CpG islands, that are GC-rich (made up of about 65% CG residues), wherein none are methylated. These are associated with the promoters of 56% of mammalian genes, including all ubiquitously expressed genes. One to two percent of the human genome are CpG clusters, and there is an inverse relationship between CpG methylation and transcriptional activity.
Protein methylation typically takes place on arginine or lysine amino acid residues in the protein sequence.[1] Arginine can be methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylated arginine) or one on both nitrogens (symmetric dimethylated arginine) by peptidylarginine methyltransferases (PRMTs). Lysine can be methylated once, twice or three times by lysine methyltransferases. Protein methylation has been most-studied in the histones. The transfer of methyl groups from S-adenosyl methionine to histones is catalyzed by enzymes known as histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.[2][3] Protein methylation is one type of post-translational modification.
Embryonic development
Main article: ReprogrammingDuring the development of germ cells their genomes are demethylated, while chromosomes in the somatic cells retain the parental methylation patterns. After that, a De novo methylation of the germ cells occurs, modifying and adding epigenetic information to the genome based on the sex of the individual.[4]
After fertilization of an oocyte and formations of a zygote, its combined genome is demethylated and remethylated again (with the exception of the imprinted genes). By blastula stage, the methylation of the embryonic cells is complete.
The process of demethylation/remethylation is referred to as "reprogramming".[5] The importance of methylation was shown in knockout mutants without DNA methyltransferase, which all died at the morula stage.[6]
Postnatal development
Increasing evidence is revealing a role of methylation in the interaction of environmental factors with genetic expression. Differences in maternal care during the first 6 days of life in the rat induce differential methylation patterns in some promoter regions, thus influencing gene expression.[7] Furthermore, even-more-dynamic processes such as interleukin signaling have been shown to be regulated by methylation.[8]
Research in humans has shown that repeated high level activation of the body’s stress system, especially in early childhood, can alter methylation processes and lead to changes in the chemistry of the individual’s DNA. The chemical changes can disable genes and prevent the brain from properly regulating its response to stress. Researchers and clinicians have drawn a link between this neurochemical disregulation and the development of chronic health problems such as depression, obesity, diabetes, hypertension, and coronary artery disease. [9][10][11][12][13]
Cancer
The pattern of methylation has recently become an important topic for research. Studies have found that in normal tissue, methylation of a gene is mainly localized to the coding region, which is CpG-poor. In contrast, the promoter region of the gene is unmethylated, despite a high density of CpG islands in the region.
Neoplasia is characterized by "methylation imbalance" where genome-wide hypomethylation is accompanied by localized hypermethylation and an increase in expression of DNA methyltransferase.[14] The overall methylation state in a cell might also be a precipitating factor in carcinogenesis as evidence suggests that genome-wide hypomethylation can lead to chromosome instability and increased mutation rates.[15] The methylation state of some genes can be used as a biomarker for tumorigenesis. For instance, hypermethylation of the pi-class glutathione S-transferase gene (GSTP1) appears to be a promising diagnostic indicator of prostate cancer.[16]
In cancer, the dynamics of genetic and epigenetic gene silencing are very different. Somatic genetic mutation leads to a block in the production of functional protein from the mutant allele. If a selective advantage is conferred to the cell, the cells expand clonally to give rise to a tumor in which all cells lack the capacity to produce protein. In contrast, epigenetically mediated gene silencing occurs gradually. It begins with a subtle decrease in transcription, fostering a decrease in protection of the CpG island from the spread of flanking heterochromatin and methylation into the island. This loss results in gradual increases of individual CpG sites, which vary between copies of the same gene in different cells.[17]
Bacterial host defense
In addition, adenosine or cytosine methylation is part of the restriction modification system of many bacteria. Bacterial DNAs are methylated periodically throughout the genome. A methylase is the enzyme that recognizes a specific sequence and methylates one of the bases in or near that sequence. Foreign DNAs (which are not methylated in this manner) that are introduced into the cell are degraded by sequence-specific restriction enzymes. Bacterial genomic DNA is not recognized by these restriction enzymes. The methylation of native DNA acts as a sort of primitive immune system, allowing the bacteria to protect themselves from infection by bacteriophage. These restriction enzymes are the basis of restriction fragment length polymorphism (RFLP) testing, used to detect DNA polymorphisms.
In chemistry
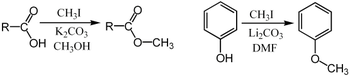
The term methylation in organic chemistry refers to the alkylation process used to describe the delivery of a CH3 group.[18] This is commonly performed using electrophilic methyl sources - iodomethane, dimethyl sulfate, dimethyl carbonate, or less commonly with the more powerful (and more dangerous) methylating reagents of methyl triflate or methyl fluorosulfonate (magic methyl), which all react via SN2 nucleophilic substitution. For example a carboxylate may be methylated on oxygen to give a methyl ester, an alkoxide salt RO− may be likewise methylated to give an ether, ROCH3, or a ketone enolate may be methylated on carbon to produce a new ketone.
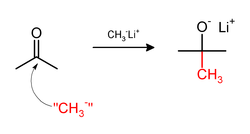
On the other hand, the methylation may involve use of nucleophilic methyl compounds such as methyllithium (CH3Li) or Grignard reagents (CH3MgX). For example, CH3Li will methylate acetone, adding across the carbonyl (C=O) to give the lithium alkoxide of tert-butanol:
Purdie methylation
Purdie methylation is a specific method for the methylation at oxygen of carbohydrates using iodomethane and silver oxide.[19]
See also
- alkylation
- Bisulfite sequencing - the biochemical method used to determine the presence or absence of methyl groups on a DNA sequence
- MethDB DNA Methylation Database
- Microscale thermophoresis - a biophysical method to determine the methylisation state of DNA[20]
External links
- deltaMasses Detection of Methylations after Mass Spectrometry
References
- ^ Christopher Walsh (2006). "Chapter 5 - Protein Methylation". Posttranslational modification of proteins: expanding nature's inventory. Roberts and Co. Publishers. ISBN 0-9747077-3-2.. http://www.roberts-publishers.com/walsh/chapter5.pdf.
- ^ Grewal SI, Rice JC (2004). "Regulation of heterochromatin by histone methylation and small RNAs". Curr. Opin. Cell Biol. 16 (3): 230–8. doi:10.1016/j.ceb.2004.04.002. PMID 15145346. http://linkinghub.elsevier.com/retrieve/pii/S0955067404000535.
- ^ Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001). "Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly". Science 292 (5514): 110–3. doi:10.1126/science.1060118. PMID 11283354. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=11283354.
- ^ Carroll, Sean B.; Wessler, Susan R.; Griffiths, Anthony J. F.; Lewontin, Richard C. (2008). Introduction to genetic analysis (9th ed.). New York: W.H. Freeman and CO. p. 403. ISBN 0-7167-6887-9.
- ^ Mann MR, Bartolomei MS (2002). "Epigenetic reprogramming in the mammalian embryo: struggle of the clones". Genome Biol. 3 (2): 1003.1–.4. doi:10.1186/gb-2002-3-2-reviews1003. PMC 139014. PMID 11864375. http://genomebiology.com/1465-6906/3/REVIEWS1003.
- ^ Woroniecki, R.; Gaikwad, A. B.; Susztak, K. (2010). "Fetal environment, epigenetics, and pediatric renal disease". Pediatric Nephrology 26 (5): 705–711. doi:10.1007/s00467-010-1714-8. PMC 3063864. PMID 21174217. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3063864.
- ^ Weaver IC (2007). "Epigenetic programming by maternal behavior and pharmacological intervention. Nature versus nurture: let's call the whole thing off". Epigenetics 2 (1): 22–8. doi:10.4161/epi.2.1.3881. PMID 17965624. http://www.landesbioscience.com/journals/epi/abstract.php?id=3881.
- ^ Bird A (2003). "Il2 transcription unleashed by active DNA demethylation". Nat. Immunol. 4 (3): 208–9. doi:10.1038/ni0303-208. PMID 12605226.
- ^ Caldji, C; Hellstrom, IC; Zhang, TY; Diorio, J; Meaney, MJ (2011). "Environmental regulation of the neural epigenome". FEBS letters. doi:10.1016/j.febslet.2011.03.032. PMID 21420958.
- ^ Champagne, FA; Curley, JP (2005). "How social experiences influence the brain". Current opinion in neurobiology 15 (6): 704–9. doi:10.1016/j.conb.2005.10.001. PMID 16260130.
- ^ Champagne, FA; Weaver, IC; Diorio, J; Dymov, S; Szyf, M; Meaney, MJ (2006). "Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring". Endocrinology 147 (6): 2909–15. doi:10.1210/en.2005-1119. PMID 16513834.
- ^ Felitti, VJ; Anda, RF; Nordenberg, D; Williamson, DF; Spitz, AM; Edwards, V; Koss, MP; Marks, JS (1998). "Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. The Adverse Childhood Experiences (ACE) Study". American journal of preventive medicine 14 (4): 245–58. PMID 9635069.
- ^ McEwen, BS; Gianaros, PJ (2010). "Central role of the brain in stress and adaptation: Links to socioeconomic status, health, and disease". Annals of the New York Academy of Sciences 1186: 190–222. doi:10.1111/j.1749-6632.2009.05331.x. PMC 2864527. PMID 20201874. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2864527.
- ^ Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP (1998). "Alterations in DNA methylation: a fundamental aspect of neoplasia". Adv. Cancer Res. 72: 141–96. doi:10.1016/S0065-230X(08)60702-2. PMID 9338076.
- ^ Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R (1998). "DNA hypomethylation leads to elevated mutation rates". Nature 395 (6697): 89–93. doi:10.1038/25779. PMID 9738504.
- ^ Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG (2004). "GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer". J. Cell. Biochem. 91 (3): 540–52. doi:10.1002/jcb.10740. PMID 14755684.
- ^ Jones PA, Baylin SB (2002). "The fundamental role of epigenetic events in cancer". Nat. Rev. Genet. 3 (6): 415–28. doi:10.1038/nrg816. PMID 12042769.
- ^ March, Jerry; Smith, Michael W. (2001). March's advanced organic chemistry: reactions, mechanisms, and structure. New York: Wiley. ISBN 0-471-58589-0.
- ^ Purdie T, Irvine JC (1903). "The alkylation of sugars". J. Chem. Soc. 83: 1021–37. doi:10.1039/CT9038301021.
- ^ Wienken CJ, Baaske P, Duhr S, Braun D (2011). "Thermophoretic melting curves quantify the conformation and stability of RNA and DNA". Nucleic Acids Research 39 (8): e52–e52. doi:10.1093/nar/gkr035. http://nar.oxfordjournals.org/content/early/2011/02/04/nar.gkr035.full?keytype=ref&ijkey=FsmS2IhsrZ8wdBr.
Protein primary structure and posttranslational modifications General N terminus C terminus Single specific AAs Phosphorylation · Sulfation · Porphyrin ring linkage · Adenylylation · Flavin linkage · Topaquinone (TPQ) formationAspartateGlutamateTransglutaminationMethylation · Acetylation · Acylation · Adenylylation · Hydroxylation · Ubiquitination · Sumoylation · ADP-ribosylation · Deamination · Oxidative deamination to aldehyde · O-glycosylation · Imine formation · Glycation · CarbamylationDiphthamide formation · AdenylylationCrosslinks between two AAs Sulfilimine bondLysine-TyrosylquinoneLysine tyrosylquinone (LTQ) formationTryptophan-TryptophylquinoneThree consecutive AAs
(Chromophore formation)4-(p-hydroxybenzylidene)-5-imidazolinone formationCrosslinks between four AAs Secondary structure→Categories:- Epigenetics
- Organic reactions
- Posttranslational modification
- Methylating agents
Wikimedia Foundation. 2010.