- Candida albicans
-
Candida albicans 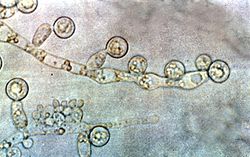
Scientific classification Kingdom: Fungi Phylum: Ascomycota Subphylum: Saccharomycotina Class: Saccharomycetes Order: Saccharomycetales Family: Saccharomycetaceae Genus: Candida Species: C. albicans Binomial name Candida albicans
(C.P. Robin)
Berkhout 1923Synonyms Candida stellatoidea[1]
Oidium albicans[2]
Candida albicans is a diploid fungus that grows both as yeast and filamentous cells and a causal agent of opportunistic oral and genital infections in humans.[3][4] Systemic fungal infections (fungemias) including those by C. albicans have emerged as important causes of morbidity and mortality in immunocompromised patients (e.g., AIDS, cancer chemotherapy, organ or bone marrow transplantation). C. albicans biofilms may form on the surface of implantable medical devices. In addition, hospital-acquired infections by C. albicans have become a cause of major health concerns.
C. albicans is commensal and a constituent of the normal gut flora comprising microorganisms that live in the human mouth and gastrointestinal tract. C. albicans lives in 80% of the human population without causing harmful effects, although overgrowth of the fungus results in candidiasis (candidosis). Candidiasis is often observed in immunocompromised individuals such as HIV-infected patients. A common form of candidiasis restricted to the mucosal membranes in mouth or vagina is thrush, which is usually easily cured in people who are not immunocompromised. For example, higher prevalence of colonization of C. albicans was reported in young individuals with tongue piercing, in comparison to non-tongue-pierced matched individuals.[5] To infect host tissue, the usual unicellular yeast-like form of C. albicans reacts to environmental cues and switches into an invasive, multicellular filamentous form, a phenomenon called dimorphism.[3]
Contents
Genome
One of the most important features of the C. albicans genome is the occurrence of numeric and structural chromosomal rearrangements as means of generating genetic diversity, named chromosome length polymorphisms (contraction/expansion of repeats), reciprocal translocations, chromosome deletions and trisomy of individual chromosomes. These karyotypic alterations lead to changes in the phenotype, which is an adaptation strategy of this fungus. These mechanisms will be better understood with the complete analysis of the C. albicans genome.
The C. albicans genome for strain SC5314 was sequenced at the Stanford DNA Sequencing and Technology Center.[6][7] The genome of the WO1 strain was sequenced by the Broad Institute of MIT and Harvard.[8]
The sequencing of the C. albicans genome and subsequently of the genomes of several other medically relevant Candida species has profoundly and irreversibly changed the way Candida species are now investigated and understood.[4] The C. albicans genome sequencing effort was launched in October 1996. Successive releases of the sequencing data and genome assemblies have occurred in the last 10 years, culminating in the release of the diploid assembly 19, which provided a haploid version of the genome along with data on allelic regions in the genome.[4] A refined assembly 20 with the eight assembled C. albicans chromosomes was released in the summer of 2006. Importantly, the availability of sequencing data prior to the completion of the genome sequence has made it possible to start C. albicans post-genomics early on. In this regard, genome databases have been made available to the research community providing different forms of genome annotation. These have been merged in a community-based annotation hosted by the Candida Genome Database. The availability of the genome sequence has paved the way for the implementation of post-genomic approaches to the study of C. albicans: macroarrays and then microarrays have been developed and used to study the C. albicans transcriptome; proteomics has also been developed and complements transcriptional analyses; furthermore, systematic approaches are becoming available to study the contribution of each C. albicans gene in different contexts. Other Candida genome sequences have been, or are being, determined: C. glabrata, C. dubliniensis, C. parapsilosis, C. guilliermondii, C. lusitaniae, and C. tropicalis. These species will soon enter the post-genomic era as well and provide interesting comparative data. The genome sequences obtained for the different Candida species along with those of non-pathogenic hemiascomycetes provide a wealth of knowledge on the evolutionary processes that shaped the hemiascomycete group, as well as those that may have contributed to the success of different Candida species as pathogens.[4]
An unusual feature of the Candida genus is that in many of its species (including C. albicans and C. tropicalis but not, for instance, C. glabrata) the CUG codon, which normally specifies leucine, specifies serine in these species; this is an unusual example of a departure from the universal genetic code (most such departures are in start codons or, for eukaryotes, mitochondrial genetic codes).[9][10][11] This alteration may help these Candida species (in some environments) by inducing a permanent stress response - a more generalized form of the heat shock response.[12]
The genome of C. albicans is highly dynamic, and this variability has been used advantageously for molecular epidemiological studies of C. albicans and population studies in this species. The genome sequence has allowed for identifying the presence of a parasexual cycle (no meiotic division) in C. albicans.[13] This parasexual cycle is under the control of mating-type loci and switching between white and opaque phenotypes. Investigating the role the mating process plays in the dynamics of the C. albicans population or in other aspects of C. albicans biology and pathogenicity will undoubtedly represent an important focus for future research.[4] A similar lack of meiosis was found in Saccharomyces cerevisiae altered to use the same genetic code as C. albicans.[14]
Dimorphism
Although often referred to as “dimorphic”, C. albicans is in fact polyphenic. When cultured in standard yeast laboratory medium C. albicans grows as ovoid “yeast” cells. However, mild environmental changes in temperature and pH can result in a morphological shift to pseudohyphal growth. Pseudohyphae share many similarities with yeast cells [15] but their role during candidiasis remains unknown. When C. albicans cells are grown in medium which mimic the physiological environment of a human host, they grow as “true” hyphae. The ability of C. albicans to form hyphae has been proposed as a virulence factor as these structures are often observed invading tissue and C. albicans strains which are unable to form hyphae are defective in causing infection.
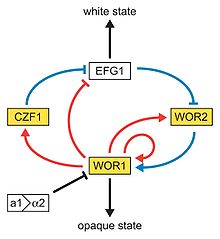 Model of the genetic network regulating the white-opaque switch. White and gold boxes represent genes enriched in the white and opaque states, respectively. The Blue lines represent relationships based on genetic epistasis. Red lines represent Wor1 control of each gene, based on Wor1 enrichment in chromatin immunoprecipitation experiments. Activation (arrowhead) and repression (bar) are inferred based on white- and opaque-state expression of each gene.
Model of the genetic network regulating the white-opaque switch. White and gold boxes represent genes enriched in the white and opaque states, respectively. The Blue lines represent relationships based on genetic epistasis. Red lines represent Wor1 control of each gene, based on Wor1 enrichment in chromatin immunoprecipitation experiments. Activation (arrowhead) and repression (bar) are inferred based on white- and opaque-state expression of each gene.
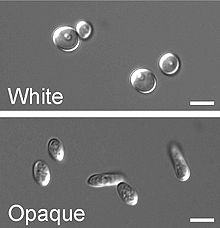
In a process that superficially resembles dimorphism, C. albicans undergoes a process called phenotypic switching, in which different cellular morphologies are generated spontaneously. One of the classically studied strains that undergoes phenotypic switching is WO-1,[16] which consists of two phases: one that grows as round cells in smooth white colonies and one that is rod-like and grows as flat gray colonies. The other strain known to undergo switching is 3153A; this strain produces at least seven different colony morphologies. In both the WO-1 and 3153A strains, the different phases convert spontaneously to the other(s) at a low frequency. The switching is reversible, and colony type can be inherited from one generation to another. While several genes that are expressed differently in different colony morphologies have been identified, some recent efforts focus on what might control these changes. Further, whether there is a potential molecular link between dimorphism and phenotypic switching is a tantalizing question.
In the 3153A strain, a gene called SIR2 (for silent information regulator) has been found that seems to be important for phenotypic switching. SIR2 was originally found in Saccharomyces cerevisiae (brewer's yeast), where it is involved in chromosomal silencing—a form of transcriptional regulation, in which regions of the genome are reversibly inactivated by changes in chromatin structure (chromatin is the complex of DNA and proteins that make chromosomes). In yeast, genes involved in the control of mating type are found in these silent regions, and SIR2 represses their expression by maintaining a silent-competent chromatin structure in this region. The discovery of a C. albicans SIR2 implicated in phenotypic switching suggests it too has silent regions controlled by SIR2, in which the phenotype-specific genes may reside.
Another potential regulatory molecule is Efg1p, a transcription factor found in the WO-1 strain that regulates dimorphism, and more recently has been suggested to help regulate phenotypic switching. Efg1p is expressed only in the white and not in the gray cell-type, and overexpression of Efg1p in the gray form causes a rapid conversion to the white form.[17][18]
So far, very few data say that dimorphism and phenotypic switching use common molecular components. However, it is not inconceivable that phenotypic switching may occur in response to some change in the environment as well as being a spontaneous event. How SIR2 itself is regulated in S. cerevisiae may yet provide clues as to the switching mechanisms of C. albicans.
Heterozygosity
The heterozygosity of the Candida genome exceeds that found in other genomes and is widespread among clinical isolates. Non-synonymous single base polymorphisms result in two proteins that differ in one or several amino acids that may confer functional differences for each protein. This situation considerably increases the number of different proteins encoded by the genome.[19]
See also
- Torula yeast (Candida utilis)
- Candidiasis
- Undecylenic acid (Castor oil derivative) for candida fungus infections.
- Leaky gut syndrome for damage to the bowel or gut (increased permeability to gut wall or bowel lining).
References
- ^ Candida albicans at NCBI Taxonomy browser, url accessed 2006-12-26
- ^ "Factors Affecting the Morphology of Candida Albicans" Dan Otho McClary Annals of the Missouri Botanical Garden, Vol. 39, No. 2 (May, 1952), pp. 137-164. doi:10.2307/2394509
- ^ a b Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 0-8385-8529-9.
- ^ a b c d e dEnfert C; Hube B (editors) (2007). Candida: Comparative and Functional Genomics. Caister Academic Press. ISBN 9781904455134.
- ^ Zadik Yehuda, Burnstein Saar, Derazne Estella, Sandler Vadim, Ianculovici Clariel, Halperin Tamar (March 2010). "Colonization of Candida: prevalence among tongue-pierced and non-pierced immunocompetent adults". Oral Dis 16 (2): 172–5. doi:10.1111/j.1601-0825.2009.01618.x. PMID 19732353.
- ^ Jones T, Federspiel NA, Chibana H, et al. (May 2004). "The diploid genome sequence of Candida albicans". Proceedings of the National Academy of Sciences of the United States of America 101 (19): 7329–34. doi:10.1073/pnas.0401648101. PMC 409918. PMID 15123810. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=15123810.
- ^ Braun BR, van Het Hoog M, d'Enfert C, et al. (July 2005). "A Human-Curated Annotation of the Candida albicans Genome". PLoS genetics 1 (1): 36–57. doi:10.1371/journal.pgen.0010001. PMC 1183520. PMID 16103911. http://www.plosgenetics.org/article/info:doi/10.1371/journal.pgen.0010001.
- ^ "Candida Database". Broad Institute. 2008-10-29. http://www.broad.mit.edu/annotation/genome/candida_albicans/MultiHome.html. Retrieved 2008-11-02.
- ^ Ohama T., et al. (August 1993). "Non-universal decoding of the leucine codon CUG in several Candida species". Nucleic Acids Research 21 (17): 1039–4045. doi:10.1093/nar/21.17.4039. PMC 309997. PMID 8371978. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=309997.
- ^ Arnaud, MB; Costanzo, MC; Inglis, DO; Skrzypek, MS; Binkley, J; Shah, P; Binkley, G; Miyasato, SR; Sherlock, G. "CGD Help: Non-standard Genetic Codes". Candida Genome Database. http://www.candidagenome.org/help/code_tables.shtml. Retrieved 30 October 2011.
- ^ Andrzej (Anjay) Elzanowski and Jim Ostell (07 July 2010). "The Alternative Yeast Nuclear Code". The Genetic Codes. Bethesda, Maryland, U.S.A.: National Center for Biotechnology Information (NCBI). http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi#SG12. Retrieved 30 October 2011.
- ^ Santos, MA; Cheesman, C; Costa, V; Moradas-Ferreira, P; Tuite, MF (February 1999). "Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp.". Molecular Microbiology 31 (3): 937-947. doi:10.1046/j.1365-2958.1999.01233.x. PMID 10048036.
- ^ Butler, G., et al. (June 4 2009). "Evolution of pathogenicity and sexual reproduction in eight Candida genomes". Nature 459 (7247): 657–662. doi:10.1038/nature08064. PMC 2834264. PMID 19465905. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2834264.
- ^ Silva, RM; Paredes, JA; Moura, GR; Manadas, B; Lima-Costa, T; Rocha, R; Miranda, I; Gomes, AC; Koerkamp, MJ; Perrot, M; Holstege, FC; Boucherie, H; Santos, MA (31 October 2007). "Critical roles for a genetic code alteration in the evolution of the genus Candida". EMBO Journal 26 (21): 4555-4565. doi:10.1038/sj.emboj.7601876. PMC 2063480. PMID 17932489. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2063480. Retrieved 31 October 2011.
- ^ Berman J, Sudbery PE (2002). "Candida Albicans: a molecular revolution built on lessons from budding yeast". Nat Rev Genet. 3 (12): 918–930. doi:10.1038/nrg948. PMID 12459722.
- ^ Rikkerrink E, Magee B, Magee P (1988). "Opaque-white phenotype transition: a programmed morphological transition in Candida albicans". J. Bact. 170 (2): 895–899. PMC 210739. PMID 2828333. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=210739.
- ^ Sonneborn A, Tebarth B, Ernst J (1999). "Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator". Infect. Immu. 67 (9): 4655–4660.
- ^ Srikantha T, Tsai L, Daniels K, Soll D (2000). "EFG1 Null Mutants of Candida albicans Switch but Cannot Express the Complete Phenotype of White-Phase Budding Cells". J. Bact. 182 (6): 1580–1591. doi:10.1128/JB.182.6.1580-1591.2000. PMC 94455. PMID 10692363. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=94455.
- ^ Larriba G; Calderone RA (2008). "Heterozygosity and Loss of Heterozygosity in Candida albicans". Pathogenic Fungi: Insights in Molecular Biology. Caister Academic Press. ISBN 978-1-904455-32-5. http://www.horizonpress.com/pat2.
Further reading
- Jones T, Federspiel NA, Chibana H, et al. (May 2004). "The diploid genome sequence of Candida albicans". Proceedings of the National Academy of Sciences 101 (19): 7329–34. doi:10.1073/pnas.0401648101. PMC 409918. PMID 15123810. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=15123810.
- Waldman A, Gilhar A, Duek L, Berdicevsky I (May 2001). "Incidence of Candida in psoriasis--a study on the fungal flora of psoriatic patients". Mycoses 44 (3–4): 77–81. doi:10.1046/j.1439-0507.2001.00608.x. PMID 11413927. http://www.blackwell-synergy.com/openurl?genre=article&sid=nlm:pubmed&issn=0933-7407&date=2001&volume=44&issue=3-4&spage=77.
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD (October 2007). "Interlocking Transcriptional Feedback Loops Control White-Opaque Switching in Candida albicans". PLoS biology 5 (10): e256. doi:10.1371/journal.pbio.0050256. PMC 1976629. PMID 17880264. http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0050256.
- Rossignol T, Lechat P, Cuomo C, Zeng Q, Moszer I, d'Enfert C (January 2008). "CandidaDB: a multi-genome database for Candida species and related Saccharomycotina". Nucleic Acids Research 36 (Database issue): D557–61. doi:10.1093/nar/gkm1010. PMC 2238939. PMID 18039716. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=18039716.
- "How Candida albicans switches phenotype - and back again: the SIR2 silencing gene has a say in Candida's colony type". NCBI Coffeebreak. 1999-11-24. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View..ShowSection&rid=coffeebrk.chapter.20. Retrieved 2008-11-02.
External links
Infectious diseases · Mycoses and Mesomycetozoea (B35–B49, 110–118) Superficial and
cutaneous
(dermatomycosis):
Tinea=skin;
Piedra (exothrix/
endothrix)=hairBy locationTinea barbae/Tinea capitis (Kerion) · Tinea corporis (Ringworm, Dermatophytid) · Tinea cruris · Tinea manuum · Tinea pedis (Athlete's foot) · Tinea unguium/Onychomycosis (White superficial onychomycosis · Distal subungual onychomycosis · Proximal subungual onychomycosis)
Tinea corporis gladiatorum · Tinea faciei · Tinea imbricata · Tinea incognito · FavusBy organismEpidermophyton floccosum · Microsporum canis · Microsporum audouinii · Trichophyton interdigitale/mentagrophytes · Trichophyton tonsurans · Trichophyton schoenleini · Trichophyton rubrumOtherHortaea werneckii (Tinea nigra) · Piedraia hortae (Black piedra)Subcutaneous,
systemic,
and opportunisticDimorphic
(yeast+mold)Coccidioides immitis/Coccidioides posadasii (Coccidioidomycosis, Disseminated coccidioidomycosis, Primary cutaneous coccidioidomycosis. Primary pulmonary coccidioidomycosis) · Histoplasma capsulatum (Histoplasmosis, Primary cutaneous histoplasmosis, Primary pulmonary histoplasmosis, Progressive disseminated histoplasmosis) · Histoplasma duboisii (African histoplasmosis) · Lacazia loboi (Lobomycosis) · Paracoccidioides brasiliensis (Paracoccidioidomycosis)OtherBlastomyces dermatitidis (Blastomycosis, North American blastomycosis, South American blastomycosis) · Sporothrix schenckii (Sporotrichosis) · Penicillium marneffei (Penicilliosis)Yeast-likeCandida albicans (Candidiasis, Oral, Esophageal, Vulvovaginal, Chronic mucocutaneous, Antibiotic candidiasis, Candidal intertrigo, Candidal onychomycosis, Candidal paronychia, Candidid, Diaper candidiasis, Congenital cutaneous candidiasis, Perianal candidiasis, Systemic candidiasis, Erosio interdigitalis blastomycetica) · C. glabrata · C. tropicalis · C. lusitaniae · Pneumocystis jirovecii (Pneumocystosis, Pneumocystis pneumonia)Mold-likeAspergillus (Aspergillosis, Aspergilloma, Allergic bronchopulmonary aspergillosis, Primary cutaneous aspergillosis) · Exophiala jeanselmei (Eumycetoma) · Fonsecaea pedrosoi/Fonsecaea compacta/Phialophora verrucosa (Chromoblastomycosis) · Geotrichum candidum (Geotrichosis) · Pseudallescheria boydii (Allescheriasis)Entomophthorales
(Entomophthoramycosis)Basidiobolus ranarum (Basidiobolomycosis) · Conidiobolus coronatus/Conidiobolus incongruus (Conidiobolomycosis)Enterocytozoon bieneusi/Encephalitozoon intestinalisMesomycetozoea Ungrouped Alternariosis · Fungal folliculitis · Fusarium (Fusariosis) · Granuloma gluteale infantum · Hyalohyphomycosis · Otomycosis · PhaeohyphomycosisCategories:- Candida
- Yeasts
- Sequenced genomes
Wikimedia Foundation. 2010.