- RNA splicing
-
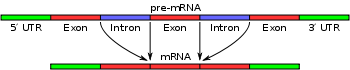
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation. For many eukaryotic introns, splicing is done in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs), but there are also self-splicing introns.
Contents
Splicing pathways
Several methods of RNA splicing occur in nature; the type of splicing depends on the structure of the spliced intron and the catalysts required for splicing to occur.
Spliceosomal introns
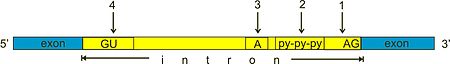
Spliceosomal introns often reside in between eukaryotic protein-coding genes. Within the intron, a 3' splice site, 5' splice site, and branch site are required for splicing. The 5' splice site or splice donor site includes an almost invariant sequence GU at the 5' end of the intron, within a larger, less highly conserved consensus region. The 3' splice site or splice acceptor site terminates the intron with an almost invariant AG sequence. Upstream (5'-ward) from the AG there is a region high in pyrimidines (C and U), or polypyrimidine tract. Upstream from the polypyrimidine tract is the branch point, which includes an adenine nucleotide.[1][2] Point mutations in the underlying DNA or errors during transcription can activate a "cryptic splice site" in part of the transcript that usually is not spliced. This results in a mature messenger RNA with a missing section of an exon. In this way a point mutation, which usually only affects a single amino acid, can manifest as a deletion in the final protein.
Spliceosome formation and activity
Splicing is catalyzed by the spliceosome which is a large RNA-protein complex composed of five small nuclear ribonucleoproteins (snRNPs, pronounced 'snurps' ). The RNA components of snRNPs interact with the intron and may be involved in catalysis. Two types of spliceosomes have been identified (the major and minor) which contain different snRNPs.
- Major
- The major spliceosome splices introns containing GU at the 5' splice site and AG at the 3' splice site. It is composed of the U1, U2, U4, U5, and U6 snRNPs and is active in the nucleus. In addition, a number of proteins including U2AF and SF1 are required for the assembly of the spliceosome.[2][3]
- E Complex-U1 binds to the GU sequence at the 5' splice site, along with accessory proteins/enzymes ASF/SF2, U2AF (binds at the Py-AG site), SF1/BBP (BBP=Branch Binding Protein);
- A Complex-U2 binds to the branch site and ATP is hydrolyzed;
- B1 Complex-U5/U4/U6 trimer binds, and the U5 binds exons at the 5' site, with U6 binding to U2;
- B2 Complex-U1 is released, U5 shifts from exon to intron and the U6 binds at the 5' splice site;
- C1 Complex-U4 is released, U6/U2 catalyzes transesterification, that make 5'end of introns ligate to the A on intron and form a lariat ,U5 binds exon at 3' splice site, and the 5' site is cleaved, resulting in the formation of the lariat;
- C2 Complex-U2/U5/U6 remain bound to the lariat, and the 3' site is cleaved and exons are ligated using ATP hydrolysis. The spliced RNA is released and the lariat debranches.
- This type of splicing is termed canonical splicing or termed the lariat pathway, which accounts for more than 99% of splicing. By contrast, when the intronic flanking sequences do not follow the GU-AG rule, noncanonical splicing is said to occur (see "minor spliceosome" below).[4]
- Minor
- The minor spliceosome is very similar to the major spliceosome, however it splices out rare introns with different splice site sequences. While the minor and major spliceosomes contain the same U5 snRNP, the minor spliceosome has different, but functionally analogous snRNPs for U1, U2, U4, and U6, which are respectively called U11, U12, U4atac, and U6atac.[5] Like the major spliceosome, it is only found in the nucleus.[6]
- Trans-splicing
- Trans-splicing is a form of splicing that joins two exons that are not within the same RNA transcript.[7]
Self-splicing
Self-splicing occurs for rare introns that form a ribozyme, performing the functions of the spliceosome by RNA alone. There are three kinds of self-splicing introns, Group I, Group II and Group III. Group I and II introns perform splicing similar to the spliceosome without requiring any protein. This similarity suggests that Group I and II introns may be evolutionarily related to the spliceosome. Self-splicing may also be very ancient, and may have existed in an RNA world present before protein. Although the two splicing mechanisms described below do not require any proteins to occur, 5 additional RNA molecules and over 50 proteins are used and hydrolyzes many ATP molecules. The splicing mechanisms use ATP in order to accurately splice mRNA's. If the cell were to not use any ATP's, the process would be highly inaccurate and many mistakes would occur.
Two transesterifications characterize the mechanism in which group I introns are spliced:
- 3'OH of a free guanine nucleoside (or one located in the intron) or a nucleotide cofactor (GMP, GDP, GTP) attacks phosphate at the 5' splice site.
- 3'OH of the 5'exon becomes a nucleophile and the second transesterification results in the joining of the two exons.
The mechanism in which group II introns are spliced (two transesterification reaction like group I introns) is as follows:
- The 2'OH of a specific adenosine in the intron attacks the 5' splice site, thereby forming the lariat
- The 3'OH of the 5' exon triggers the second transesterification at the 3' splice site thereby joining the exons together.
tRNA splicing
tRNA (also tRNA-like) splicing is another rare form of splicing that usually occurs in tRNA. The splicing reaction involves a different biochemistry than the spliceomsomal and self-splicing pathways. Ribonucleases cleave the RNA and ligases join the exons together.
Evolution
Splicing occurs in all the kingdoms or domains of life, however, the extent and types of splicing can be very different between the major divisions. Eukaryotes splice many protein-coding messenger RNAs and some non-coding RNAs. Prokaryotes, on the other hand, splice rarely and mostly non-coding RNAs. Another important difference between these two groups of organisms is that prokaryotes completely lack the spliceosomal pathway.
Because spliceosomal introns are not conserved in all species, there is debate concerning when spliceosomal splicing evolved. Two models have been proposed: the intron late and intron early models (see intron evolution).
Splicing Diversity Eukaryotes Prokaryotes Spliceosomal + - Self-splicing + + tRNA + + Biochemical mechanism
Spliceosomal splicing and self-splicing involves a two-step biochemical process. Both steps involve transesterification reactions that occur between RNA nucleotides. tRNA splicing, however, is an exception and does not occur by transesterification.
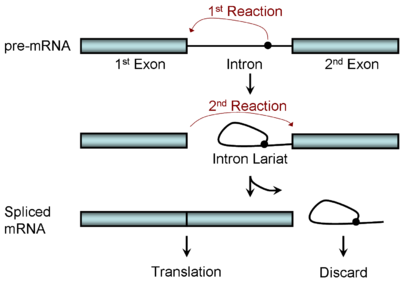
Spliceosomal and self-splicing transesterification reactions occur via two sequential transesterification reactions. First, the 2'OH of a specific branch-point nucleotide within the intron that is defined during spliceosome assembly performs a nucleophilic attack on the first nucleotide of the intron at the 5' splice site forming the lariat intermediate. Second, the 3'OH of the released 5' exon then performs a nucleophilic attack at the last nucleotide of the intron at the 3' splice site thus joining the exons and releasing the intron lariat.
Alternative splicing
In many cases, the splicing process can create a range of unique proteins by varying the exon composition of the same messenger RNA. This phenomenon is then called alternative splicing. Alternative splicing can occur in many ways. Exons can be extended or skipped, or introns can be retained.
Experimental manipulation of splicing
Splicing events can be experimentally altered[8] by binding steric-blocking antisense oligos such as Morpholinos or Peptide nucleic acids to snRNP binding sites, to the branchpoint nucleotide that closes the lariat,[9] or to splice-regulatory element binding sites.[10]
Splicing errors
Common errors:
- Mutation of a splice site resulting in loss of function of that site. Results in exposure of a premature stop codon, loss of an exon, or inclusion of an intron.
- Mutation of a splice site reducing specificity. May result in variation in the splice location, causing insertion or deletion of amino acids, or most likely, a disruption of the reading frame.
- Displacement of a splice site, leading to inclusion or exclusion of more RNA than expected, resulting in longer or shorter exons.
Many splicing errors are safeguarded by a cellular quality control mechanism termed Nonsense-mediated mRNA decay [NMD].[11]
Protein splicing
In addition to RNA, proteins can undergo splicing. Although the biomolecular mechanisms are different, the principle is the same: parts of the protein, called inteins instead of introns, are removed. The remaining parts, called exteins instead of exons, are fused together. Protein splicing has been observed in a wide range of organisms, including bacteria, archaea, plants, yeast and humans.[12]
See also
- cDNA
- Exon
- Intron
- Primary transcript
- Spliceosome
- Minor spliceosome
- Exon Junction Complex
References
- ^ Clancy, Suzanne (2008). "RNA Splicing: Introns, Exons and Spliceosome". Nature Education 1 (1). http://www.nature.com/scitable/topicpage/rna-splicing-introns-exons-and-spliceosome-12375. Retrieved 31 March 2011.
- ^ a b Black, Douglas L. (2003). "Mechanisms of alternative pre-messenger RNA splicing". Annual Reviews of Biochemistry 72 (1): 291–336. doi:10.1146/annurev.biochem.72.121801.161720. PMID 12626338.
- ^ Matlin, AJ; Clark F, Smith, CWJ (May 2005). "Understanding alternative splicing: towards a cellular code". Nature Reviews 6 (5): 386–398. doi:10.1038/nrm1645. PMID 15956978.
- ^ Ng B, Yang F, Huston DP, et al. (December 2004). "Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes". J. Allergy Clin. Immunol. 114 (6): 1463–70. doi:10.1016/j.jaci.2004.09.006. PMID 15577853.
- ^ Patel AA, Steitz JA (2003). "Splicing double: insights from the second spliceosome". Nat. Rev. Mol. Cell Biol. 4 (12): 960–70. doi:10.1038/nrm1259. PMID 14685174.
- ^ Friend K, Kolev NG, Shu MD, Steitz JA (2008). "Minor-class splicing occurs in the nucleus of the Xenopus oocyte". RNA 14 (8): 1459–62. doi:10.1261/rna.1119708. PMC 2491479. PMID 18567814. http://rnajournal.cshlp.org/content/14/8/1459.full.
- ^ Di Segni G, Gastaldi S, Tocchini-Valentini GP (May 2008). "Cis- and trans-splicing of mRNAs mediated by tRNA sequences in eukaryotic cells". Proc. Natl. Acad. Sci. U.S.A. 105 (19): 6864–9. doi:10.1073/pnas.0800420105. PMC 2383978. PMID 18458335. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2383978.
- ^ Draper BW, Morcos PA, Kimmel CB (2001). "Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown". Genesis 30 (3): 154–6. doi:10.1002/gene.1053. PMID 11477696.
Sazani P, Kang SH, Maier MA, et al. (1 October 2001). "Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs". Nucleic Acids Res. 29 (19): 3965–74. PMC 60237. PMID 11574678. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=60237. - ^ Morcos, PA (2007). "Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos.". Biochem. Biophys. Res. Commun. 358 (2): 521–7. doi:10.1016/j.bbrc.2007.04.172. PMID 17493584.
- ^ Bruno IG, Jin W, Cote GJ (2004-10-15). "Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements". Hum. Mol. Genet. 13 (20): 2409–20. doi:10.1093/hmg/ddh272. PMID 15333583. http://hmg.oxfordjournals.org/cgi/content/full/13/20/2409.(Epub August 27, 2004)
- ^ Danckwardt S, Neu-Yilik G, Thermann R, Frede U, Hentze MW, Kulozik AE (2002). "Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay". Blood 99 (5): 1811–6. doi:10.1182/blood.V99.5.1811. PMID 11861299. http://bloodjournal.hematologylibrary.org/cgi/content/full/99/5/1811.
- ^ Ken-ichi Hanada, James C. Yang (2005). "Increased Novel biochemistry: post-translational protein splicing and other lessons from the school of antigen processing" (PDF). J Mol Med 83 (6): 420–428. doi:10.1007/s00109-005-0652-6. PMID 15759099. http://www.springerlink.com/content/tu4144hm703k5713/fulltext.pdf.
External links
Gene expression Introduction to genetics Transcription (Transcription factors, RNA Polymerase,promoter) Prokaryotic / Archaeal / Eukaryotic
post-transcriptional modification (hnRNA,5' capping,Splicing,Polyadenylation)Translation (Ribosome,tRNA) Prokaryotic / Archaeal / Eukaryotic
post-translational modification (functional groups, peptides, structural changes)Gene regulation Nuclear Precursor mRNA · 5' cap formation · Polyadenylation (CPSF, CstF, PAP, PAB2, CFI, CFII) · Poly(A)-binding protein
RNA splicing: intron/exon · snRNP · spliceosome (minor spliceosome, U1) · alternative splicing · pre-mRNA processing factor (PLRG1, PRPF3, PRPF4, PRPF4B, PRPF6, PRPF8, PRPF18, PRPF19, PRPF31, PRPF38A, PRPF38B, PRPF39, PRPF40A, PRPF40B)
RNA editing · PolyuridylationCytosolic Categories:
Wikimedia Foundation. 2010.