- Enhancer (genetics)
-
In genetics, an enhancer is a short region of DNA that can be bound with proteins (namely, the trans-acting factors, much like a set of transcription factors) to enhance transcription levels of genes (hence the name) in a gene cluster. While enhancers are usually cis-acting, an enhancer does not need to be particularly close to the genes it acts on, and sometimes need not be located on the same chromosome.[1]
Contents
Enhancer locations
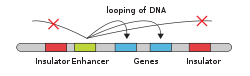
In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so that although the enhancer DNA is far from the gene in regard to the number of nucleotides, it is geometrically close to the promoter and gene. This allows it to interact with the general transcription factors and RNA polymerase II.[citation needed]
An enhancer may be located upstream or downstream of the gene it regulates. Furthermore, an enhancer need not be located near to the transcription initiation site to affect transcription, as some have been found located in several hundred thousand base pairs upstream or downstream of the start site. Enhancers do not act on the promoter region itself, but are bound by activator proteins. These activator proteins interact with the mediator complex, which recruits polymerase II and the general transcription factors which then begin transcribing the genes. Enhancers can also be found within introns. An enhancer's orientation may even be reversed without affecting its function. Additionally, an enhancer may be excised and inserted elsewhere in the chromosome, and still affect gene transcription. That is one reason that intron polymorphisms may have effects although they are not translated.[citation needed]
Theories
Currently, there are two different theories on the information processing that occurs on enhancers:[2]
- Enhanceosomes - rely on highly cooperative, coordinated action and can be disabled by single point mutations that move or remove the binding sites of individual proteins.[citation needed]
- Flexible billboards - less integrative, multiple proteins independently regulate gene expression and their sum is read in by the basal transcriptional machinery[citation needed]
HACNS1
HACNS1 (also known as CENTG2 and located in the Human Accelerated Region 2) is a gene enhancer "that may have contributed to the evolution of the uniquely opposable human thumb, and possibly also modifications in the ankle or foot that allow humans to walk on two legs". Evidence to date shows that of the 110,000 gene enhancer sequences identified in the human genome, HACNS1 has undergone the most change during the evolution of humans following the split with the ancestors of chimpanzees.[3]
GADD45G
An enhancer near the gene GADD45g has been described that may regulate brain growth in chimpanzees and other mammals, but not in humans.[4][5] The GADD45G regulator in mice and chimps is active in regions of the brain where cells that form the cortex, ventral forebrain, and thalamus are located and may suppress further neurogenesis. Loss of the GADD45G enhancer in humans may contribute to an increase of certain neuronal populations and to forebrain expansion in humans.[citation needed]
Enhancers in Developmental Biology
The development, differentiation and growth of cells and tissues require precisely regulated patterns of gene expression. Enhancers work as cis-regulatory elements to mediate both spatial and temporal control of development by turning on transcription in specific cells and/or repressing it in other cells. Thus, the particular combination of transcription factors and other DNA-binding proteins in a developing tissue controls which genes will be expressed in that tissue. Enhancers allow the same gene to be used in diverse processes in space and time.[citation needed]
Identification and Characterization
Enhancers can be identified by enhancer trap techniques using a reporter gene or by comparative sequence analysis and computational genomics. In genetically tractable models such as the fruit fly Drosophila melanogaster, for example, a reporter construct such as the lacZ gene can be randomly integrated into the genome using a P element transposon. If the reporter gene integrates near an enhancer, its expression will reflect the expression pattern driven by that enhancer. Thus, staining the flies for LacZ expression or activity and cloning the sequence surrounding the integration site can identify the enhancer sequence.[6]
In the comparative genomics approach, sequence conservation of non-coding regions can be indicative of enhancers. Sequences from multiple species are aligned, and conserved regions are identified computationally.[7] Identified sequences can then be attached to a reporter gene such green fluorescent protein or lacZ to determine the in vivo pattern of gene expression produced by the enhancer when injected into an embryo. mRNA expression of the reporter can be visualized by in situ hybridization, which provides a more direct measure of enhancer activity, since it is not subjected to the complexities of translation and protein folding. Although much evidence has pointed to sequence conservation for critical developmental enhancers, other work has shown that the function of enhancers can be conserved with little or no primary sequence conservation. For example, the RET enhancers in humans have very little sequence conservation to those in zebrafish, yet both species’ sequences produce nearly identical patterns of reporter gene expression in zebrafish.[7] Additionally, algorithms such as TFSEARCH or JASPAR can be used to identify putative transcription factor binding sites to further characterize an enhancer sequence.[citation needed]
Enhancers in Invertebrate Segmentation
The enhancers determining early segmentation in Drosophila melanogaster embryos are among the best characterized developmental enhancers. In the early fly embryo, the gap gene transcription factors are responsible for activating and repressing a number of segmentation genes, such as the pair rule genes. The gap genes are expressed in blocks along the anterior-posterior axis of the fly along with other maternal effect transcription factors, thus creating zones within which different combinations of transcription factors are expressed. The pair-rule genes are separated from one another by non-expressing cells. Moreover, the stripes of expression for different pair-rule genes are offset by a few cell diameters from one another. Thus, unique combinations of pair-rule gene expression create spatial domains along the anterior-posterior axis to set up each of the 14 individual segments. The 480 bp enhancer responsible for driving the sharp stripe two of the pair-rule gene even-skipped (eve) has been well-characterized. The enhancer contains 12 different binding sites for maternal and gap gene transcription factors. Activating and repressing sites overlap in sequence. Eve is only expressed in a narrow stripe of cells that contain high concentrations of the activators and low concentration of the repressors for this enhancer sequence. Other enhancer regions drive eve expression in 6 other stripes in the embryo.[8]
Enhancers in Vertebrate Patterning
Establishing body axes is a critical step in animal development. During mouse embryonic development, Nodal, a transforming growth factor-beta superfamily ligand, is a key gene involved in patterning both the anterior-posterior axis and the left-right axis of the early embryo. The Nodal gene contains two enhancers: the Proximal Epiblast Enhancer (PEE) and the Asymmetric Enhancer (ASE). The PEE is upstream of the Nodal gene and drives Nodal expression in the portion of the primitive streak that will differentiate into the node (also referred to as the primitive node).[9] The PEE turns on Nodal expression in response to a combination of Wnt signaling plus a second, unknown signal; thus, a member of the LEF/TCF transcription factor family likely binds to a TCF binding site in the cells in the node. Diffusion of Nodal away from the node forms a gradient which then patterns the extending anterior-posterior axis of the embryo.[10] The ASE is an intronic enhancer bound by the fork head domain transcription factor Fox1. Early in development, Fox1-driven Nodal expression establishes the visceral endoderm. Later in development, Fox1 binding to the ASE drives Nodal expression on the left side of the lateral plate mesoderm, thus establishing left-right asymmetry necessary for asymmetric organ development in the mesoderm.[11]
Establishing three germ layers during gastrulation is another critical step in animal development. Each of the three germ layers has unique patterns of gene expression that promote their differentiation and development. The endoderm is specified early in development by Gata4 expression, and Gata4 goes on to direct gut morphogenesis later. Gata4 expression is controlled in the early embryo by an intronic enhancer that binds another forkhead domain transcription factor, FoxA2. Initially the enhancer drives broad gene expression throughout the embryo, but the expression quickly becomes restricted to the endoderm, suggesting that other repressors may be involved in its restriction. Late in development, the same enhancer restricts expression to the tissues that will become the stomach and pancreas. An additional enhancer is responsible for maintaining Gata4 expression in the endoderm during the intermediate stages of gut development.[12]
Multiple enhancers promote developmental robustness
Some genes involved in critical developmental processes contain multiple enhancers of overlapping function. Secondary enhancers, or “shadow enhancers,” may be found many kilobases away from the primary enhancer (“primary” usually refers to the first enhancer discovered, which is often closer to the gene it regulates). On its own, each enhancer drives nearly identical patterns of gene expression. Are the two enhancers truly redundant? Recent work has shown that multiple enhancers allow fruit flies to survive environmental perturbations, such as an increase in temperature. When raised at an elevated temperature, a single enhancer sometimes fails to drive the complete pattern of expression, whereas the presence of both enhancers permits normal gene expression.[13]
Enhancers and the evolution of developmental mechanisms
One theme of research in evolutionary developmental biology (“evo-devo”) is investigating the role of enhancers and other cis-regulatory elements in producing morphological changes via developmental differences between species.[citation needed]
Stickleback PitX1
Recent work has investigated the role of enhancers in morphological changes in threespine stickleback fish. Sticklebacks exist in both marine and freshwater environments, but sticklebacks in many freshwater populations have completely lost their pelvic fins (appendages homologous to the posterior limb of tetrapods). Pitx1 is a homeobox gene involved in posterior limb development in vertebrates. Preliminary genetic analyses indicated that changes in the expression of this gene were responsible for pelvic reduction in sticklebacks. Fish expressing only the freshwater allele of Pitx1 do not have pelvic spines, whereas fish expressing a marine allele retain pelvic spines. A more thorough characterization showed that a 500 base pair enhancer sequence is responsible for turning on Pitx1 expression in the posterior fin bud. This enhancer is located near a chromosomal fragile site--a sequence of DNA that is likely to be broken and thus more likely to be mutated as a result of imprecise DNA repair. This fragile site has caused repeated, independent losses of the enhancer responsible for driving PitX1 expression in the pelvic spines in isolated freshwater population, and without this enhancer, freshwater fish fail to develop pelvic spines.[14]
Enhancers in Drosophila wing pattern evolution
Pigmentation patterns provide one of the most striking and easily scored differences between different species of animals. Pigmentation of the Drosophila wing has proven to be a particularly amenable system for studying the development of complex pigmentation phenotypes. The Drosophila guttifera wing has 12 dark pigmentation spots and 4 lighter gray intervein patches. Pigment spots arise from expression of the yellow gene, whose product produces black melanin. Recent work has shown that two enhancers in the yellow gene produce gene expression in precisely this pattern – the vein spot enhancer drives reporter gene expression in the 12 spots, and the intervein shade enhancer drives reporter expression in the 4 distinct patches. These two enhancers are responsive to the Wnt signaling pathway, which is activated by ‘’wingless’’ expression at all of the pigmented locations. Thus, in the evolution of the complex pigmentation phenotype, the yellow pigment gene evolved enhancers responsive to the wingless signal and wingless expression evolved at new locations to produce novel wing patterns.[15]
References
- ^ Spilianakis, Charalampos G.; Lalioti, Maria D.; Town, Terrence; Lee, Gap Ryol; Flavell, Richard A. (2005). "Interchromosomal associations between alternatively expressed loci". Nature 435 (7042): 637–45. doi:10.1038/nature03574. PMID 15880101.
- ^ Arnosti, David N.; Kulkarni, Meghana M. (2005). "Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards?". Journal of Cellular Biochemistry 94 (5): 890–8. doi:10.1002/jcb.20352. PMID 15696541. http://www.bch.msu.edu/faculty/arnosti/Arnosti&Kulkarni2005JCB.pdf.
- ^ HACNS1: Gene enhancer in evolution of human opposable thumb[self-published source?]
- ^ McLean, Cory Y.; Reno, Philip L.; Pollen, Alex A.; Bassan, Abraham I.; Capellini, Terence D.; Guenther, Catherine; Indjeian, Vahan B.; Lim, Xinhong et al. (2011). "Human-specific loss of regulatory DNA and the evolution of human-specific traits". Nature 471 (7337): 216–9. doi:10.1038/nature09774. PMC 3071156. PMID 21390129. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3071156.
- ^ New Scientist, 12 March 2011, pages 3, 6-7[verification needed]
- ^ Hartenstein, Volker; Jan, Yuh Nung (1992). "Studying Drosophila embryogenesis with P-lacZ enhancer trap lines". Roux's Archives of Developmental Biology 201 (4): 194–220. doi:10.1007/BF00188752.
- ^ a b Visel, Axel; Bristow, James; Pennacchio, Len A. (2007). "Enhancer identification through comparative genomics". Seminars in Cell & Developmental Biology 18: 140–152. doi:10.1016/j.semcdb.2006.12.014.
- ^ Borok, M. J.; Tran, D. A.; Ho, M. C. W.; Drewell, R. A. (2009). "Dissecting the regulatory switches of development: Lessons from enhancer evolution in Drosophila". Development 137 (1): 5–13. doi:10.1242/dev.036160. PMC 2796927. PMID 20023155. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2796927.
- ^ Norris, D. P.; Robertson, E. J. (1999). "Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements". Genes & Development 13 (12): 1575–88. doi:10.1101/gad.13.12.1575. PMC 316799. PMID 10385626. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=316799.
- ^ Granier, Céline; Gurchenkov, Vasily; Perea-Gomez, Aitana; Camus, Anne; Ott, Sascha; Papanayotou, Costis; Iranzo, Julian; Moreau, Anne et al. (2011). "Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo". Developmental Biology 349 (2): 350–62. doi:10.1016/j.ydbio.2010.10.036. PMID 21047506.
- ^ Norris, DP; Brennan, J; Bikoff, EK; Robertson, EJ (2002). "The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo". Development 129 (14): 3455–68. PMID 12091315.
- ^ Rojas, Anabel; Schachterle, William; Xu, Shan-Mei; Martín, Franz; Black, Brian L. (2010). "Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer". Developmental Biology 346 (2): 346–55. doi:10.1016/j.ydbio.2010.07.032. PMC 2945415. PMID 20692247. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2945415.
- ^ Perry, Michael W.; Boettiger, Alistair N.; Bothma, Jacques P.; Levine, Michael (2010). "Shadow Enhancers Foster Robustness of Drosophila Gastrulation". Current Biology 20 (17): 1562–1567. doi:10.1016/j.cub.2010.07.043. PMID 20797865.
- ^ Chan, Y. F.; Marks, M. E.; Jones, F. C.; Villarreal, G.; Shapiro, M. D.; Brady, S. D.; Southwick, A. M.; Absher, D. M. et al. (2009). "Adaptive Evolution of Pelvic Reduction in Sticklebacks by Recurrent Deletion of a Pitx1 Enhancer". Science 327 (5963): 302–305. doi:10.1126/science.1182213. PMC 3109066. PMID 20007865. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3109066.
- ^ Werner, Thomas; Koshikawa, Shigeyuki; Williams, Thomas M.; Carroll, Sean B. (2010). "Generation of a novel wing colour pattern by the Wingless morphogen". Nature 464 (7292): 1143–1148. doi:10.1038/nature08896. PMID 20376004.
External links
Transcriptional regulation prokaryoticeukaryoticbothPromotion Promoter (Pribnow box, TATA box, BRE, CAAT box, Response element) · Enhancer (E-box, Response element) · Insulator · SilencerInitiation (prokaryotic,
eukaryotic)Transcription start siteElongation Termination
(prokaryotic,
eukaryotic)Categories:
Wikimedia Foundation. 2010.