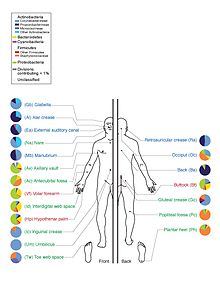
- Skin flora
-
The skin flora are the microorganisms which reside on the skin. Most research has been upon those that reside upon the 2 square metres of human skin. Many of them are bacteria of which there are around 1000 species upon human skin from 19 phyla.[1][2] The total number of bacteria on an average human has been estimated at 1012 (1,000,000,000,000).[3] Most are found in the superficial layers of the epidermis and the upper parts of hair follicles.
Skin flora are usually non-pathogenic, and either commensals (are not harmful to their host) or mutualistic (offer a benefit). The benefits bacteria can offer include preventing transient pathogenic organisms from colonizing the skin surface, either by competing for nutrients, secreting chemicals against them, or stimulating the skin's immune system.[4] However, resident microbes can cause skin diseases and enter the blood system creating life-threatening diseases particularly in immunosuppressed people.[4] Hygiene to control such flora is important in preventing the transmission of antibiotic resistant hospital-acquired infections.
A major nonhuman skin flora is Batrachochytrium dendrobatidis, a chytrid and non-hyphal zoosporic fungus that causes chytridiomycosis, an infectious disease thought to be responsible for the decline in amphibian populations.
Contents
Species variety
Bacteria
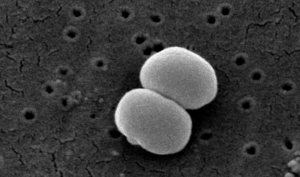 Scanning electron microscope image of Staphylococcus epidermidis one of roughly a thousand bacteria species present on human skin. Though usually not pathogenic, it can cause skin infections and even life threatening illnesses in those that are immunocompromised.
Scanning electron microscope image of Staphylococcus epidermidis one of roughly a thousand bacteria species present on human skin. Though usually not pathogenic, it can cause skin infections and even life threatening illnesses in those that are immunocompromised.
The estimate of the number of species present on skin bacteria has been radically changed by the use of 16S ribosomal RNA to identify bacterial species present on skin samples direct from their genetic material. Previously such identification had depended upon microbiological culture upon which many varieties of bacteria did not grow and so were hidden to science.[1]
Staphylococcus epidermidis and Staphylococcus aureus were thought from cultural based research to be dominant. However 16S ribosomal RNA research finds that while common, these species make up only 5% of skin bacteria.[5] However, skin variety provides a rich and diverse habitat for bacteria. Most come from four phyla: Actinobacteria (51.8%), Firmicutes (24.4%), Proteobacteria (16.5%), and Bacteroidetes (6.3%).
There are three main ecological areas: moist, dry and sebaceous. Propionibacteria and Staphylococci species were the main species in sebaceous areas. In moist places on the body Corynebacteria together with Staphylococci dominate. In dry areas, there is a mixture of species but b-Proteobacteria and Flavobacteriales are dominant. Ecologically, sebaceous areas had greater species richness than moist and dry one. The areas with least similarity between people in species were the spaces between fingers, the spaces between toes, axillae, and umbilical cord stump. Most similarly were beside the nostril, nares (inside the nostril), and on the back.[1]
Frequency of the best studied skin microbes[4] Organism observations Staphylococcus epidermidis Common, occasionally pathogenic Staphylococcus aureus Infrequent, usually pathogenic Staphylococcus warneri Infrequent, occasionally pathogenic Streptococcus pyogenes Infrequent, usually pathogenic Streptococcus mitis Frequent, occasionally pathogenic Propionibacterium acnes Frequent, occasionally pathogenic Corynebacterium spp. Frequent, occasionally pathogenic Acinetobacter johnsonii Frequent, occasionally pathogenic Pseudomonas aeruginosa Infrequent, occasionally pathogenic Fungal
A study of the area between toes in 100 young adults found 14 different genera of fungi. These include yeasts such as Candida albicans, Rhodotorula rubra, Torulopsis and Trichosporon cutaneum, dermatophytes (skin living fungi) such as Microsporum gypseum, and Trichophyton rubrum and nondermatophyte fungi (opportunistic fungi that can live in skin) such as Rhizopus stolonifer, Trichosporon cutaneum, Fusarium, Scopulariopsis brevicaulis, Curvularia, Alternaria alternata, Paecilomyces, Aspergillus flavus and Penicillium species.[6]
Relationship to host
Skin microflora can be commensals, mutualistic or pathogens. Often they can be all three depending upon the strength of the person's immune system.[4] Research upon the immune system in the gut and lungs has shown that microflora aids immunity development: however such research has only started upon whether this is the case with the skin.[4] Pseudomonas aeruginosa is an example of a mutualistic bacterium that can turn into a pathogen and cause disease: if it gains entry into the blood system it can result in infections in bone, joint, gastrointestinal, and respiratory systems. It can also cause dermatitis. However, Pseudomonas aeruginosa produces antimicrobial substances such as pseudomonic acid that are exploited commercially such as Mupirocin. This works against staphylococcal and streptococcal infections. Pseudomonas aeruginosa also produces substances that inhibit the growth of fungus species such as Candida krusei, Candida albicans, Torulopsis glabrata, Saccharomyces cerevisiae and Aspergillus fumigatus.[7] It can also inhibit the growth of Helicobacter pylori.[8] So important is its antimicrobial actions that it has been noted that "removing P. aeruginosa from the skin, through use of oral or topical antibiotics, may inversely allow for aberrant yeast colonization and infection."[4]
Another aspect of bacteria is the generation of body odor. Sweat is odorless but Propionibacteria in adolescent and adult sebaceous glands can turn its amino acids into propionic acid. Staphylococcus epidermidis create the other source of body odor: isovaleric acid (3-methyl butanoic acid).[9] In addition to these, strong foot odor is due to Bacillus subtilis.[10]
Skin defenses
Antimicrobial peptides
The skin creates antimicrobial peptides such as cathelicidins that control the proliferation of skin microbes. Cathelicidins not only reduce microbe numbers directly but also cause the secretion of cytokine release which induces inflammation, angiogenesis, and reepithelialization. Conditions such as atopic dermatitis have been linked to the suppression in cathelicidin production. In rosacea abnormal processing of cathelicidin cause inflammation. Psoriasis has been linked to self-DNA created from cathelicidin peptides that causes autoinflammation. A major factor controlling cathelicidin is vitamin D3.[11]
Acidity
The superficial layers of the skin are naturally acidic (pH 4-4.5) due to lactic acid in sweat and produced by skin bacteria.[12] At this pH mutualistic flora such as Staphylococci, Micrococci, Corynebacterium and Propionibacteria grow but not transient bacteria such as Gram negative bacteria like Escherichia and Pseudomonas or Gram positive ones such as Staphylococcus aureus or Candida albicans.[12] Another factor effecting the growth of pathological bacteria is that the antimicrobial substances secreted by the skin are enhanced in acidic conditions.[12] In alkaline conditions, bacteria cease to be attached to the skin and are more readily shed. It has been observed that the skin also swells under alkaline conditions and opens up allowing move to the surface.[12]
Immune system
If activated, the immune system in the skin produces cell-mediated immunity against microbes such as dermatophytes (skin fungi).[13] One reaction is to increase stratum corneum turnover and so shed the fungus from the skin surface. Skin fungi such as Trichophyton rubrum have evolved to create substances that limit the immune response to them.[13] The shedding of skin is a general means to control the build up flora upon the skin surface.
Clinical
Skin diseases
Microorganisms play a role in noninfectious skin diseases such as atopic dermatitis,[14] rosacea, psoriasis,[15] and acne[16] Damaged skin can cause nonpathogenic bacteria to become pathogenic.[17]
Infected devices
Skin microbes are source of infected medical devices such as catheters.[18]
Hygiene
Contagion
Skin flora do not readily pass between people: 30 seconds of moderate friction and dry hand contact results in a transfer of only 0.07% of natural hand flora from naked with a greater percentage from gloves.[19]
Removal
The most effective (60 to 80% reduction) antimicrobial washing is with ethanol, isopropanol, and n-propanol. Viruses are most affected by high (95%) concentrations of ethanol, while bacteria are more affected by n-propanol.[20]
Unmedicated soaps are not very effective as illustrated by the following data. Health care workers washed their hands once in nonmedicated liquid soap for 30 seconds. The students/technicians for 20 times.[21]
Skin flora upon two hospital groups in colony-forming units per mL. group and hand skin condition unwashed washed Health care workers healthy 3.47 3.15 Health care workers damaged 3.33 3.29 Students/technicians healthy 4.39 3.54 Students/technicians damaged 4.58 4.43 An important use of hand washing is to prevent the transmission of antibiotic resistant skin flora that cause hospital-acquired infections such as Methicillin-resistant Staphylococcus aureus. While such flora have become antibiotic resistant due to antibiotics there is no evidence that recommended antiseptics or disinfectants selects for antibiotic-resistant organisms when used in hand washing.[22] However, many strains of organisms are resistant to some of the substances used in antibacterial soaps such as Triclosan.[22]
One survey of bar soaps in dentist clinics found they all had their own flora and on average from two to five different genera of microorganisms with those used most more likely to have more species varieties.[23] Another survey of bar soaps in public toilets found even more flora.[24] Another study found that very dry soaps are not infected while all are that rest in pools of water.[25] However, research upon soap that was specially infected found that soap flora do not transmit to the hands.[26]
Damaged skin
Washing skin repeatedly can damage the protective external layer and cause transepidermal loss of water. This can be seen in roughness characterized by scaling and dryness, itchiness, dermatitis provoked by microorganisms and allergens penetrating the corneal layer and redness. Wearing gloves can cause further problems since it produces a humid environment favoring the growth of microbes and also contains irritants such as latex and talcum powder.[27]
Hand washing can damage skin because the stratum corneum top layer of skin consists of 15 to 20 layers of keratin disks, corneocytes, each of which is each surrounded by a thin film of skin lipids which can be removed by alcohols and detergents.[28]
Damaged skin defined by extensive cracking of skin surface, widespread reddening or occasional bleeding has also been found to be more frequently colonized by Staphylococcus hominis and these were more likely to methicillin resistant.[27] Though not related to greater antibiotic resistance, damaged skin was also more like to be colonized by Staphylococcus aureus, gram-negative bacteria, Enterococci and Candida.[27]
Comparison with other flora
The skin flora is different from that of the gut which is predominately Firmicutes and Bacteroidetes.[29] There is also low level of variation between people that is not found in gut studies.[5] Both gut and skin flora however lack the diversity found in soil flora.[1]
See also
- Bacterial disease
- Body odor
- Gut flora
- Human flora
- Human microbiome project
- Medical microbiology
- Microbial ecology
- Microflora
- Oral microbiology
- Skin
- Vaginal flora
References
- ^ a b c d Grice EA, Kong HH, Conlan S. (2009). Topographical and Temporal Diversity of the Human Skin Microbiome, Science, 324: 1190 - 1192. doi:10.1126/science.1171700 PMID 19478181
- ^ Pappas S. (2009). Your Body Is a Wonderland ... of Bacteria. ScienceNOW Daily News
- ^ Todar K. Normal Bacterial Flora of Humans Todar's Online Textbook of Bacteriology.
- ^ a b c d e f Cogen AL, Nizet V, Gallo RL. (2008). Skin microbiota: a source of disease or defence? Br J Dermatol. 158(3):442-55. PMID 18275522
- ^ a b Grice EA, Kong HH, Renaud G, Young AC; NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA. (2008). A diversity profile of the human skin microbiota. Genome Res. 18(7):1043-50. PMID 18502944
- ^ Oyeka CA, Ugwu LO. (2002). Fungal flora of human toe webs. Mycoses. 45(11-12):488-91. PMID 12472726
- ^ Kerr JR. (1994). Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J Clin Microbiol. 32(2):525-7. PMID 8150966
- ^ Krausse R, Piening K, Ullmann U. (2005). Inhibitory effects of various micro-organisms on the growth of Helicobacter pylori. Lett Appl Microbiol. 40(1):81-6. PMID 15613007
- ^ Ara K, Hama M, Akiba S, et al. (2006). "Foot odor due to microbial metabolism and its control". Can. J. Microbiol. 52 (4): 357–64. doi:10.1139/w05-130. PMID 16699586.
- ^ Ara K, Hama M, Akiba S, Koike K, Okisaka K, Hagura T, Kamiya T, Tomita F. (2006). Foot odor due to microbial metabolism and its control. Can J Microbiol. 52(4):357-64. PMID 16699586
- ^ Schauber J, Gallo RL. (2008). Antimicrobial peptides and the skin immune defense system. J Allergy Clin Immunol. 122(2):261-6. PMID 18439663
- ^ a b c d Lambers H, Piessens S, Bloem A, Pronk H, Finkel P. (2006). Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int J Cosmet Sci. 28(5):359-70. PMID 18489300
- ^ a b Dahl MV. (1993).Suppression of immunity and inflammation by products produced by dermatophytes. J Am Acad Dermatol. 28(5 Pt 1):S19-S23. PMID 8496406
- ^ Baker BS. (2006). The role of microorganisms in atopic dermatitis. Clin Exp Immunol. 144(1):1-9. PMID 16542358
- ^ Paulino LC, Tseng CH, Strober BE, Blaser MJ. (2006). Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 44(8):2933-41. PMID 16891514
- ^ Holland KT, Cunliffe WJ, Roberts CD. (1977). Acne vulgaris: an investigation into the number of anaerobic diphtheroids and members of the Micrococcaceae in normal and acne skin.Br J Dermatol. 96(6):623-6. PMID 141301
- ^ Roth RR, James WD. (1988). Microbial ecology of the skin. Annu Rev Microbiol. 42:441-64. doi:10.1146/annurev.mi.42.100188.002301 PMID 3144238
- ^ Martín-Rabadán P, Gijón P, Alcalá L, Rodríguez-Créixems M, Alvarado N, Bouza E. (2008), Propionibacterium acnes is a common colonizer of intravascular catheters. J Infect. 56(4):257-60. PMID 18336916
- ^ Lingaas E, Fagernes M. (2009). Development of a method to measure bacterial transfer from hands. J Hosp Infect. 72(1):43-9. PMID 19282052
- ^ Kampf G, Kramer A. (2004). Epidemiologic background of hand hygiene and evaluation of the most important agents for scrubs and rubs. Clin Microbiol Rev. 17(4):863-93, doi:10.1128/CMR.17.4.863-893.2004 PMID 15489352
- ^ de Almeida e Borges LF, Silva BL, Gontijo Filho PP. (2007). Hand washing: changes in the skin flora. Am J Infect Control. 35(6):417-20. PMID 17660014
- ^ a b Weber DJ, Rutala WA (2006). "Use of germicides in the home and the healthcare setting: is there a relationship between germicide use and antibiotic resistance?". Infect Control Hosp Epidemiol 27 (10): 1107–19. doi:10.1086/507964. PMID 17006819.
- ^ Hegde PP, Andrade AT, Bhat K. (2006). Microbial contamination of "in use" bar soap in dental clinics. Indian J Dent Res. 17(2):70-3. PMID 17051871
- ^ Kabara JJ, Brady MB. (1984). Contamination of bar soaps under "in-use" conditions. J Environ Pathol Toxicol Oncol. 5(4-5):1-14. PMID 6394740
- ^ Afolabi BA, Oduyebo OO, Ogunsola FT. (2007). Bacterial flora of commonly used soaps in three hospitals in Nigeria. East Afr Med J. 84(10):489-95. PMID18232270
- ^ Heinze JE, Yackovich F. (1988). Washing with contaminated bar soap is unlikely to transfer bacteria. Epidemiol Infect. 101(1):135-42. PMID 3402545
- ^ a b c Larson EL, Hughes CA, Pyrek JD, Sparks SM, Cagatay EU, Bartkus JM. (1998).Changes in bacterial flora associated with skin damage on hands of health care personnel. Am J Infect Control. 26(5):513-21. PMID 9795681
- ^ Kownatzki E. (2003). Hand hygiene and skin health. J Hosp Infect. 55(4):239-45. PMID 14629966
- ^ Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. (2005). Diversity of the human intestinal microbial flora. Science. 308(5728):1635-8. doi:10.1126/science.1110591 PMID 15831718
External links
Categories:- Bacteriology
- Environmental microbiology
- Health
- Microbiology terms
- Microbiology
- Skin
Wikimedia Foundation. 2010.